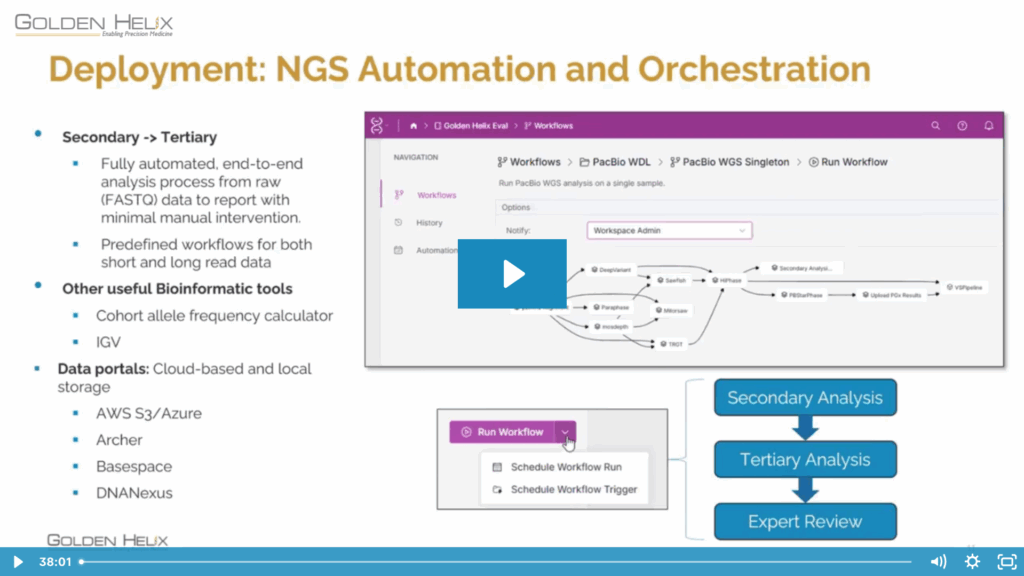
As Next-Generation Sequencing (NGS) becomes central to modern clinical diagnostics, laboratories are managing more data than ever before, and turning that data into actionable insight requires both precision and scalability. In our recent webcast, Next-Gen Sequencing at Scale: Transforming Per-Sample Outcomes into Institutional Knowledge with VSWarehouse, we explored how Golden Helix’s software ecosystem streamlines NGS analysis, bridging the gap between single-sample workflows in VarSeq and organization-wide intelligence through VSWarehouse.
From Raw Data to Clinical Report
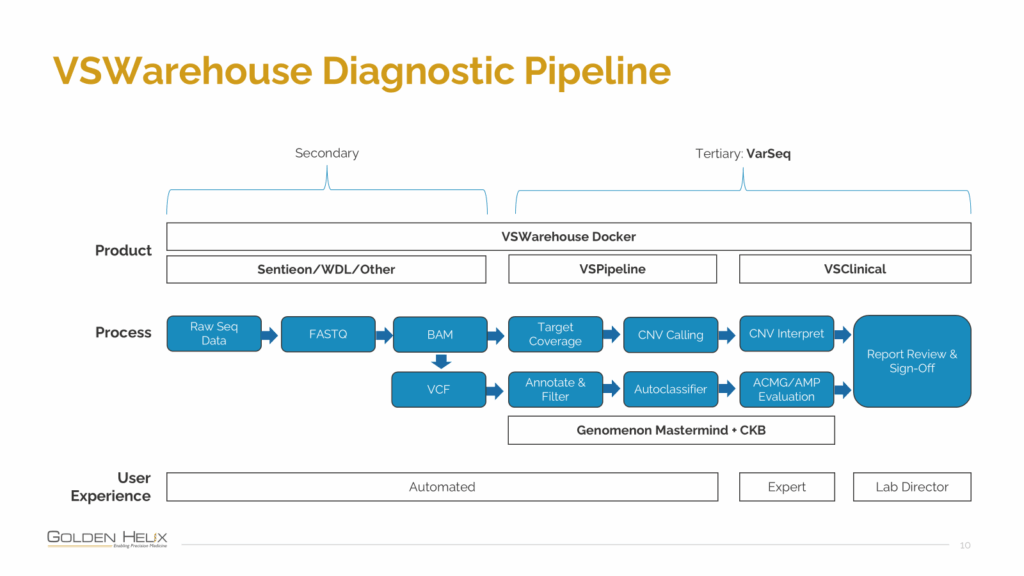
The webcast began by revisiting the VSWarehouse Diagnostic Pipeline (seen in Figure 1.), which unifies the complete secondary and tertiary analysis workflow.
Starting from raw sequencing data, the pipeline moves seamlessly through Sentieon/WDL for alignment, VSPipeline for annotation and filtering, and VSClinical for interpretation and reporting, all within a single orchestrated environment.
(Image: VSWarehouse Diagnostic Pipeline)
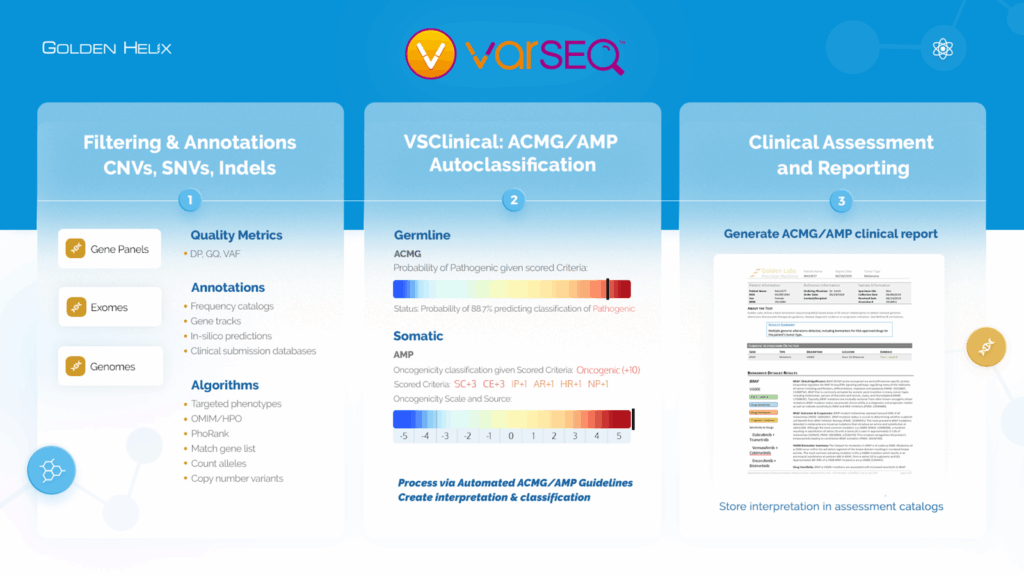
The Power of the VarSeq Workflow
VarSeq provides analysts with an intuitive, three-stage workflow (Figure 2.):
- Import and Annotate variants using extensive public and internal annotations.
- Interpret and Classify variants under ACMG/AMP guidelines using VSClinical’s automated classification engine.
- Generate Clinical Reports that summarize findings while preserving all evidence in assessment catalogs for future reference.
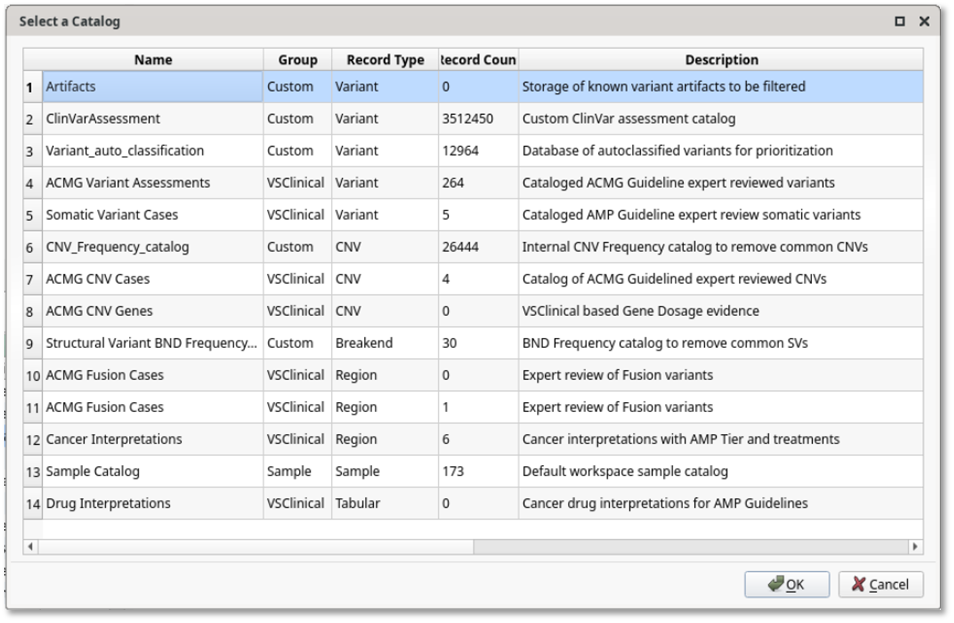
In addition to clinical reporting, assessment catalogs (Figure 3) serve as structured repositories that record every evaluated variant, ensuring prior evidence can be reused to accelerate interpretation and maintain consistency across the lab. Catalogs can also capture variant frequencies, gene dosage information, and clinical metadata, forming the foundation for institutional knowledge.
Scaling Knowledge with VSWarehouse
While VarSeq enables precise single-sample interpretation, VSWarehouse extends that precision across the entire organization.
VSWarehouse automates catalog management and variant ingestion, allowing laboratories to process thousands of samples simultaneously through parameterized workflows and Saved Export schemas.
Built on a robust server infrastructure, VSWarehouse supports multi-user collaboration and integrates with external systems through its API, transforming isolated catalog data into a shared, searchable knowledgebase.
From Workflow to Wisdom
Together, VarSeq and VSWarehouse deliver a powerful synergy:
- VarSeq ensures accuracy at the individual sample level.
- VSWarehouse scales that accuracy across cohorts and institutions, automatically enriching every future analysis with historical context.
Each new genome analyzed strengthens the organization’s collective knowledge base, reducing interpretation time, improving consistency, and driving the evolution of precision medicine at scale.
Watch the Webcast
Missed the live session? You can watch the full presentation on demand to see real-world examples of how VSWarehouse transforms routine variant analysis into institutional intelligence, or reach out to our team for more information.