Applying AMP Guidelines to Analyze Somatic Variants Today, I am thrilled to share with you the launch of a brand new eBook titled “Clinical Variant Analysis for Cancer – Applying AMP Guidelines to Analyze Somatic Variants”. We would happy to send you a complimentary copy which can be requested on our website here. The clinical utilization of Next-Gen Sequencing data… Read more »
Yesterday, I released a new version of my eBook, “Genetic Testing for Cancer – Third Edition”. We would be happy to send you one! To download a complimentary copy, please submit a request on our site here. In 1914, the German cytologist Theodor Boveri coined the phrase “Cancer is a disease of the genome.” At this time, his ideas were… Read more »
Our “Processing Hereditary Cancer Panels in VarSeq” webcast was a great lesson for viewers to learn more about the functionality of our software. If you didn’t have a chance to join us for the live event, you can watch the recording on our site here. Q: What is the best place to get clinical interpretations of breast cancer variant interpretations?… Read more »
CIViC The Clinical Interpretations of Variants in Cancer, better known as CIViC, is an open access open source, community-driven web resource available to all VarSeq users. Nature Genetics published an article that states, “CIViC accepts public knowledge contributions but requires that experts review these submissions”. Fundamentally, the focus behind CIViC is to make sure the variants contained in the database… Read more »
VarSeq enables breakthrough discoveries in cancer diagnostics by supporting gene panel testing and whole exome and genome analysis. We wanted to share our Cancer Gene Panel tutorial which covers a basic gene panel workflow with an emphasis on adding, modifying and manipulating filter chains. This tutorial will start with creating a new project from an empty project template, importing data, creating… Read more »
While clinical assessments of germline mutations have been collected in ClinVar under the stewardship of the NCBI and the collaborate effort of many testing labs, the same type of resource has been missing for mutations that could informal clinical care in Cancer. Or at least, that is what I thought until I started to work with CIViC. With the stewardship of… Read more »
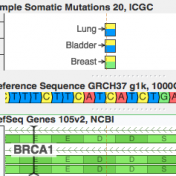
ICGC’s database is now available For quite a while, COSMIC has been synonymous with the catalog of “known somatic mutations”. It is the ClinVar of cancer mutations and invests heavily in “expert curation” (having human experts read papers and pull out references to published somatic mutations). But it turns out there is a new kid on the block, and he… Read more »
In 1914, the German cytologist Theodor Boveri coined the phrase “Cancer is a disease of the genome”. At this time, his ideas were as equally revolutionary as they were highly contested. Fast forward. More than a hundred years later, Next-Generation Sequencing effectively permits a highly sensitive analysis of cancer cells. It can help us to understand mutations associated with cancer… Read more »
Wednesday, March 2nd 12:00 pm EST Clinical labs must have the ability to go from a collection of samples and associated variants to a professional report documenting a short list of clinically relevant variants. Cancer Gene Panels are a common clinical application for genetic tests. In this webcast we will show how VarSeq and VSReports can be used to go… Read more »
Last week we conducted a webcast on “Cancer Gene Panels”. We had some excellent questions which we answered during the webcast and a few more that we didn’t get to in the allotted time. Please find answers to those questions here: 1. Are Cancer Gene Panels just another stepping stone on the way to whole exome/genome analysis? Cancer gene panels answer… Read more »
In 1914 the German cytologist Theodor Boveri coined the phrase “Cancer is a disease of the genome”. At this time his ideas were equally revolutionary as they were highly contested. Fast forward. More than hundred years later, Next-Generation Sequencing effectively permits a highly sensitive analysis of cancer cells. It can help us to understand mutations associated with cancer development and… Read more »
Dr. Folefac Aminkeng is a Postdoctoral Fellow at The Centre for Molecular Medicine and Therapeutics (CMMT) at the University of British Columbia in Vancouver, BC, Canada. He utilizes GWAS studies to identify single-nucleotide polymorphisms (SNPs) that might be associated with serious adverse drug reactions (ADRs) in cancer therapeutics. The field of pharmacogenomics—how one’s genetic makeup affects drug response—has grown exponentially… Read more »
As you may have guessed from the title of this post, we’ve had a lot of customers publishing in the first six weeks of 2011. We are always excited to hear about about our customers’ findings and how they were able to use SNP & Variation Suite to accelerate their research. (All abstracts below.) First, in the pharma world, congrats… Read more »
Recognition this month begins with Eric Londin at Coriell Institute for Medical Research for his publication in PLoS ONE: “CoAIMs: A Cost-Effective Panel of Ancestry Informative Markers for Determining Continental Origins.” (Abstract below). Also recently published in PLoS ONE is Chiara Magri with Brescia University School of Medicine on her study locating new CNVs in schizophrenia. (Abstract below) Skipping over… Read more »
We just concluded a defining year for Golden Helix. 2025 was about execution at scale. With major platform releases, national-level adoption, and meaningful progress across pharmacogenomics, oncology, and long-read sequencing, we strengthened our position as a trusted clinical genomics platform for laboratories operating in regulated, high-throughput environments. Below, we reflect on the key accomplishments of 2025 and outline our focus… Read more »
Thank you to everyone who joined our recent webcast, “VSWarehouse for Genome Centers: Scalable, Secure Whole-Genome Infrastructure for Modern Sequencing Programs,” presented by Gabe Rudy on December 10, 2025. We appreciate the strong attendance and the excellent questions regarding the operational and security needs of modern genome centers. For those who missed it or need a recap, the session focused… Read more »
As long-read sequencing solidifies its role in clinical and translational research, labs are increasingly seeking streamlined methods to transition from raw data to actionable results. With the release of VSWarehouse 3.0, Golden Helix now supports a fully integrated implementation of PacBio’s somatic workflow, providing users with a powerful, centralized environment for managing large datasets, running best-practice pipelines, and automating tertiary… Read more »
Golden Helix is excited to exhibit at the Association for Molecular Pathology (AMP) 2025 Annual Meeting in Boston, Massachusetts, where we’ll showcase our newest advancements in variant interpretation, enterprise genomics, and scalable software deployment. Visit us at Booth #543 to explore how VSWarehouse and our Bring Your Own Cloud (BYOC) solution enable laboratories to deploy secure, scalable, and compliant cloud-based… Read more »
At Golden Helix, we’re proud to see how researchers worldwide continue using our software to accelerate discovery and improve clinical decision-making. These October 2025 publications highlight how VarSeq is advancing genetic research across cancer, hereditary disease, and molecular diagnostics. From identifying new variant mechanisms to expanding our understanding of genotype–phenotype relationships, these studies demonstrate how Golden Helix tools empower teams… Read more »
Recently, Golden Helix attended the ASHG 2025 conference, where we had the opportunity to showcase our partnership with Genomenon. This collaboration took center stage at both our booth and Genomenon’s CoLab session, Automating Genomic Workflows: Cutting Interpretation Time, Accelerating Turnaround, and Increasing Diagnostic Yield. Our discussions centered on a problem that every clinical lab faces as variant volumes continue to… Read more »