ExAC CNVs were released publicly with a recent publication, providing the full set of rare CNVs called on ~60K human exomes.
While there are many public CNV databases out there, this is the first one that was derived from exome data, and thus includes both extremely rare and very small CNV events.
With the recent release of Golden Helix’s CNV calling algorithm on NGS target panel data, the availability of NGS-based CNV calls on public control samples is all the more relevant.
So we went ahead and curated the public CNV calls and they are now available as an annotation source from VarSeq, SVS and of course GenomeBrowse.
Called with XHMM on Exome Coverage Data
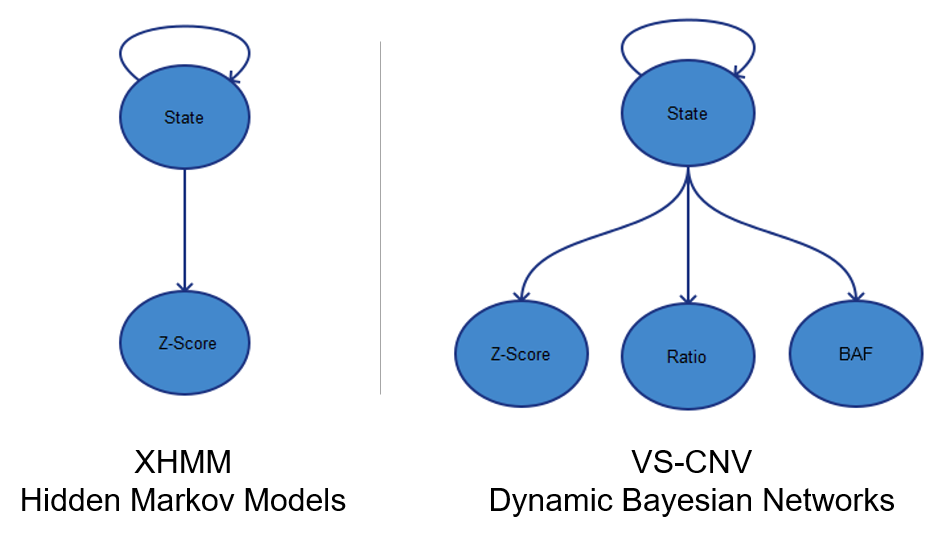
The ExAC team used XHMM for calling CNVs on their exome data. We researched this method when developing this algorithm and I can see how it would make sense on a large-cohort study such as this. XHMM uses PCA for coverage normalization, which can require some fine tuning to ensure that the correct number of principle components are selected for correction. Getting this parameter right is critical to reducing enough noise but not loosing signal. It also requires quite a few samples for the normalization to work.
In comparison, our CNV calling method does reference-sample matched control normalization, which ends up working even with as few as 10 (but recommended 30) reference samples.
Next XHMM uses Hidden Markov Models to call CNVs based on Z-scores computed on a per-target level. This was one of the most promising techniques we saw in the literature and definitely is able to pick out large and clean events with high confidence. We extended this probabilistic model in our method to not only look at Z-score, but also ratios and the allele frequency of variants in the target region.
The CNV calls provided by XHMM come with PHRED scale likelihood scores representing the confidence of the XHMM call. Only CNVs scores > 60 are included in the track. When interpreting this track, you may still want to consider calls with scores < 80 with some suspicion.
Looking at the called made by XHMM, there are a few small (one or two target) events, but the majority are larger events where the confidence score can be higher. Some genes, which are tricky to call correctly like PTEN, have no called CNVs (likely because calls were not high confidence.)
What’s in the Track
Each feature in the track represents a CNV call from a single individual. The track colors deletion calls (which may have been called as heterozygous or homozygous deletions) as red and duplications as blue.
| Type | # of Calls |
| Deletions | 49409 |
| Duplications | 77363 |
Along with the score, the reference population and the individual that the CNV was called for is provided. This allows you to get a sense of how applicable the particular call may be in the population of the sample you are studying. You can also use this field to filter down to CNVs that would be relevant to interpreting your sample using the GenomeBrowse filter control.
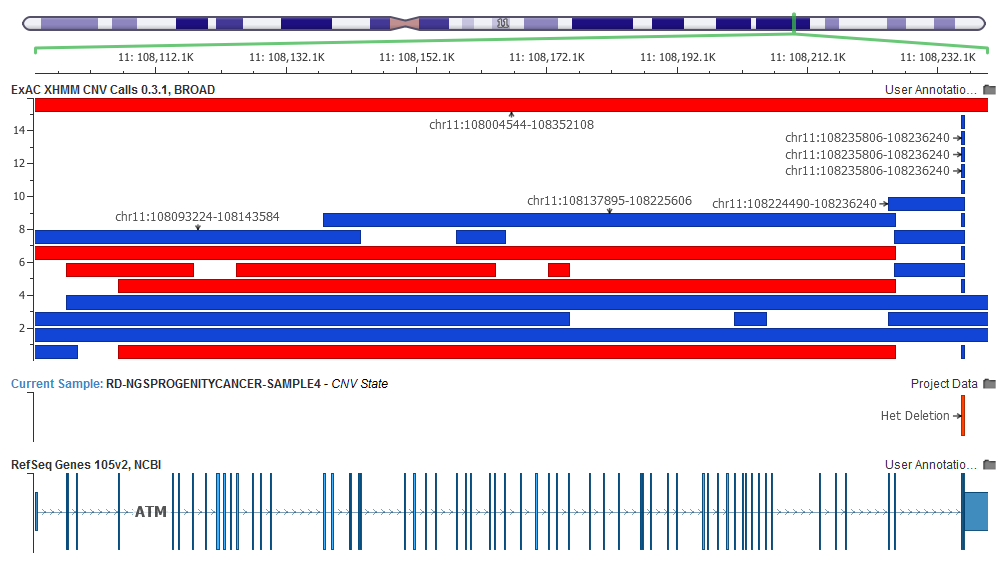
A two-target heterozygous deletion in our sample overlaps a common duplication that occurs in the ExAC 60K exomes.
Overall, the track gives you a very quick visual guide to how frequent CNV calls are (and of what type), as well as providing a fantastic complement to the visualizations of the CNVs called by VarSeq!