…so we added some automation to your automation so you can automate while you automate! Automation has been a hot topic recently and for all the right reasons. As we (proudly) watch our customers increase their sample and data volume, we are constantly seeking to provide tools to reduce click rate and optimize throughput. Furthermore, with all of the new… Read more »
Configuring a bioinformatic pipeline to reliably process genomic data is no small task. Doing so in an efficient, consistent way is an even grander challenge. Luckily, the VarSeq software suite provides a comprehensive toolbox for automation and integration. One of the first questions a new VarSeq user might ask is where processed data needs to end up. The versatility of… Read more »
A new VSPipeline command, set_data_folder_path, designed to bolster consistent input usage. By introducing this innovative command, we aim to empower users with improved data organization, flexibility, and standardization for their clinical cases and analyses. Embracing this command will not only support reproducibility but also ensure accountability, ultimately paving the way for better-informed patient care decisions. Managing Annotations and References in… Read more »
Orphanet is a public database available in VarSeq that aims to improve the current understanding and treatment options for patients affected by rare diseases. This resource was established in France in 1997 and has gradually grown to cover a consortium of over 40 countries in Europe. The primary goal of this database is to provide a universal nomenclature to classify… Read more »
VarSeq recently received major upgrades in a wide range of areas, one of these areas includes adding annotations such as GnomAD. This includes new fundamental methods of CNV ACMG guideline processing but also a large number of small additions in annotations. One addition is the application of gnomADs – Gene Constraints. This provides various metrics for pathogenicity on a per… Read more »
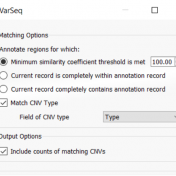
In continuation of our blog posts focusing on new features of VarSeq v2.2.2, here we will discuss the Latest Sample Assessment algorithm for both single nucleotide variants (SNVs) and copy number variants (CNVS). This algorithm annotates the variants of the project with the latest assessment from your variant catalog, which will show the history of interpretations made for the variants… Read more »
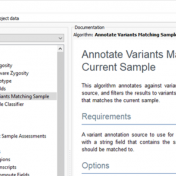
Those of you who have been attending our recent webcasts have learned about our upcoming VarSeq release. A part of that release will be an additional algorithm that will annotate variants matching the current sample. If you are not familiar with these webcasts, here are several on-demand webcasts I recommend to get you familiar with these new features: Evaluating Copy… Read more »
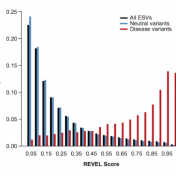
Typically, researchers are looking for rare variants in their next generation sequencing datasets. However, most of the nonsynonymous variants have unknown significance because there is an inherent difficulty in validating large numbers of rare variants or even detecting rare variants with high statistical power. In lieu of this issue, computational tools are needed as they accurately predict the pathogenicity of… Read more »
In the 1990s the genetic industry voiced a request for a variant catalog that incorporates associated variant information such as phenotypic and metabolic pathways. The call was answered by NCBI, which created dbSNP; dbSNP became publicly available in 1998 and around 1.5 million variants. Fast forward to the present and dbSNP now contains over 2 billion SNPs spanning human, rat,… Read more »
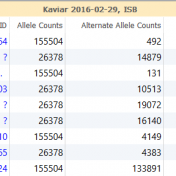
In the search for disease causing mutations it is important to determine if the variant has been previously observed in humans and at what frequency. With the advent of increasing genomic information, there is now a variety of different databases and annotation sources that can be utilized. For some, this could be a tedious task that leads only to implementing… Read more »
Relating human phenotypes to genotypes is the name of the game with OMIM, and as their website says, “is intended for use primarily by physicians and other professionals concerned with genetic disorders, by genetics researchers, and by advanced students in science and medicine.” The Online Mendelian Inheritance in Man (or OMIM) was originally created by Dr. Victor A. McKusick in… Read more »
There are many good reasons why the pursuit of the highest quality genomic interpretation would lead you to the latest human reference. It is more complete and fixes incorrect or partially missing genes that have known implications for human disease. While most major projects cataloging human populations have plans to re-do all their genomic alignments to the new human reference… Read more »
In the new Genotype Imputation tool that is coming soon to SVS, allele encoding is an important part of matching data between the target and the reference panels. If the same platform provider is being used, then A/B encoding can be used. However, it’s better to use the Reference/Alternate allele encoding associated with AGCT format to ensure accuracy. If an… Read more »
One of the tools at the top of the toolbox for researchers working with microarray data is genotype imputation. Genotype imputation is the process of inferring the genotype of one or more markers based on the correlation pattern (aka linkage disequilibrium or LD) of the surrounding markers for which genotypes are known. We have now integrated a natively ported version of BEAGLE into Golden… Read more »
We have just released SVS version 8.4.2, and included in the release is a new script for visualizing Meta-Analysis results with a Forest Plot. You can find full details on all the new and updated features included with the update in our Release Notes. Release Notes from all our software products can be found on our Support Bulletin web page… Read more »
In recent months we have been updating our public annotation library to include the most recent versions of existing sources, as well as include new sources. All of these annotation sources are compatible with our three major products, VarSeq, SVS, and GenomeBrowse, and can be used for visualization, annotation, and filtering. dbNSFP NS Functional Predictions 2.8, GHI and dbNSFP Predictions… Read more »
Golden Helix is proud to announce the release of the Golden Helix GenomeBrowse Plugin for Ion Torrent server. The new plug-in enables adding selected BAM files from Torrent Server reports directly into GenomeBrowse. The BAM files remain on the torrent server and are streamed from the server on demand using your credentials. This feature allows GenomeBrowse users to visualize genomic… Read more »
We are excited to let you know about two new scripts to aid in Numeric Regression analysis. Don’t forget about the Technical Support Bulletins which keep you up-to-date on all the latest script news. You can stream this feed via an RSS reader, receive email updates, or see the latest on the SVS splash screen. Linear and Logistic Regression with… Read more »
In a recent blog post (Comparing BEAGLE, IMPUTE2, and Minimac Imputation Methods for Accuracy, Computation Time, and Memory Usage), Autumn Laughbaum compared three imputation programs. Data can be exported from, or imported into, SVS in the standard file formats for these and other imputation programs. The goal of this blog post will be to review the different tools available to… Read more »
We are excited to let you know about new scripts to aid in filtering rows/columns and identifying unique values in a column, as well as two updated scripts. Don’t forget about the Technical Support Bulletins which keep you up-to-date on all the latest script news. You can stream this feed via an RSS reader, receive email updates, or see the… Read more »