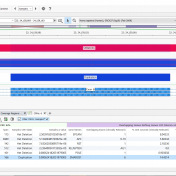
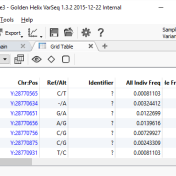
Introduction: Malignant Rhabdoid tumors (MRT) are among the most aggressive and lethal forms of infant and child cancer (1). These tumors are characterized by an unusual combination of mixed cellular elements similar to but not typical of teratomas and can originate at any anatomic location. When MRTs are present in the brain, they are called atypical teratoid/rhabdoid tumors (AT/RT), which… Read more »
Wednesday, September 21st @ 12:00 pm EDT Every day, the trove of genomic data is growing. Clinics are sequencing targeted genes at high read depths to report out genetic tests. Research groups are adding new exomes and genomes to their disease-specific cohorts. Agricultural breeders are genotyping their herds and flocks by the thousands of thousands. The conventional attitude to big… Read more »
When the new human reference genome was released over two years ago, it was hailed as a significant step forward for next generation sequencing. Compared to GRCh37, the new GRCH38 reference assembly fixed gaps, repaired incorrect sequences and offered access to sections of the genome that had been previously unaccounted for. Despite these improvements, adoption of the new assembly has… Read more »
Scaling is in our DNA: Making Genomics Accessible One of the things I absolutely love about the work we do at Golden Helix is keeping up with the changes in data analysis driven by the iterative and generational leaps in technology. But one thing has always been a constant since day one: we break preconceived notions of what scale of… Read more »
Yesterday, it was my pleasure to share in a live webcast our integrated solution for genetic data warehousing, VSWarehouse. Although we had a great set of questions at the end of our presentation, we didn’t have time to answer all of them, so here is a selection of the remaining representative questions and my answers. We can provide a hosted version… Read more »
This last week I had the pleasure of attending the fourth annual Clinical Genome Conference (TCGC) in Japantown, San Francisco and kicking off the conference by teaching a short course on Personal Genomics Variant Analysis and Interpretation. Some highlights of the conference from my perspective: Talking about clinical genomics is no longer a wonder-fest of individual case studies, but a… Read more »
If you’ve seen the recent webinars given by Gabe Rudy and Bryce Christensen, you’ve no doubt been impressed by the capabilities of VarSeq when it comes to annotation and filtering. However, we sometimes forget that the power that enables all this complex analysis can also be used in more mundane tasks like VCF subsetting. And although these day-to-day tasks don’t… Read more »
I’m sitting in the Smithsonian Air and Space Museum basking in the incredible product of human innovation and the hard work of countless engineers. My volunteer tour guide started us off at the Wright brother’s fliers and made a point of saying it was only 65 years from lift off at Kitty Hawk to the landing of a man on the moon…. Read more »
You probably haven’t spent much time thinking about how we represent genes in a genomic reference sequence context. And by genes, I really mean transcripts since genes are just a collection of transcripts that produce the same product. But in fact, there is more complexity here than you ever really wanted to know about. Andrew Jesaitis covered some of this… Read more »
Up until a few weeks ago, I thought variant classification was basically a solved problem. I mean, how hard can it be? We look at variants all the time and say things like, “Well that one is probably not too detrimental since it’s a 3 base insertion, but this frameshift is worth looking into.” What we fail to recognize is… Read more »
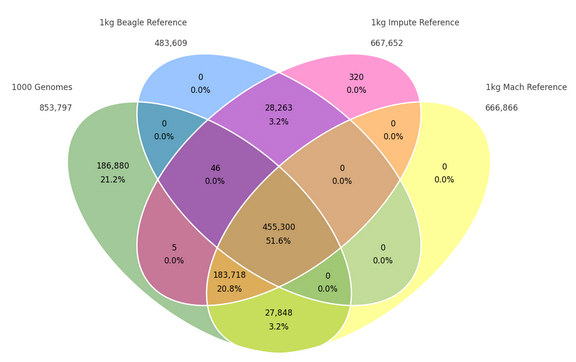
Genotype imputation is a common and useful practice that allows GWAS researchers to analyze untyped SNPs without the cost of genotyping millions of additional SNPs. In the Services Department at Golden Helix, we often perform imputation on client data, and we have our own software preferences for a variety of reasons. However, other imputation software packages have their own advantages… Read more »
When researchers realized they needed a way to report genetic variants in scientific literature using a consistent format, the Human Genome Variation Society (HGVS) mutation nomenclature was developed and quickly became the standard method for describing sequence variations. Increasingly, HGVS nomenclature is being used to describe variants in genetic variant databases as well. There are some practical issues that researchers… Read more »
I’m a believer in the signal. Whole genomes and exomes have lots of signal. Man, is it cool to look at a pile-up and see a mutation as clear as day that you arrived at after filtering through hundreds of thousands or even millions of candidates. When these signals sit right in the genomic “sweet spot” of mappable regions with… Read more »
Editor’s Note by Gabe Rudy: I’ve had the chance to exchange thoughts, emails, and blog post comments for a while now with Jeff as he has written posts on NGS Leaders and engaged with me on 23andMe. He has also worked with Golden Helix software as part of Dr. Todd Lencz’s research efforts at Zucker Hillside Hospital until he recently… Read more »
Type 2 Diabetes, Rheumatoid Arthritis, Obesity, Chrohn’s Diseases and Coronary Heart Disease are examples of common, chronic diseases that have a significant genetic component. It should be no surprise that these diseases have been the target of much genetic research. Yet over the past decade, the tools of our research efforts have failed to unravel the complete biological architecture of… Read more »
The prevalence of open-source bioinformatic tools in the genetic research space is enormous. According to The North Shore LIJ Research Institute, there are over 500 genetic analysis software packages – the great majority of which are free – as of August 2010. Open-source tools are incredibly important in genetics. They allow new methodologies to be created and expanded. They tie… Read more »
I’m a huge supporter of the Free and Open Source Software movement. I’ve written more about R than anything else on my blog, all the code I post on my blog is free and open-source, and a while back I invited you to steal my blog under a cc-by-sa license. Every now and then, however, something comes along that just might be worth paying… Read more »
Ever since the release of SNP & Variation Suite (SVS) 7.4 back in January, our software engineers have been hard at work developing new functionality for the next version, to enable researchers to have even more control over their data. (Okay, well really, they started working on it before 7.4 even came out.) While the full details of 7.5 are… Read more »
In a series of previous blog posts, I gave an overview of Next Generation Sequencing trends and technologies. In Part Two of the three part series, the range of steps and programs used in the bioinfomatics of NGS data was defined as primary, secondary and tertiary analysis. In Part Three I went into more details on the needs and workflows… Read more »
A GPU can produce an enormous boost in performance for many scientific computing applications. Since we announced the availability of SNP & Variation Suite’s incorporation of GPUs to dramatically speed up copy number segmentation, we’ve received numerous inquiries on recommendations for what GPU to purchase. Unfortunately the technical terminology and choices can be a bit confusing. In this article I… Read more »