The recent release of VarSeq 2.6.0 was filled with so many customer-requested features (for example, our long-awaited PGx workflow!) that some of our other new features have not yet had their time in the spotlight. For this blog, we are thrilled to announce that with the release of VarSeq 2.6.0, we have made WGS CNV calling more user-friendly than ever… Read more »
The release of VarSeq version 2.6.0 provides many new features. Most notably is our support for tertiary analysis of Pharmacogenomic (PGx) data. VarSeq not only calls the necessary gene diplotypes for your PGx panels but also handles large batches of samples from the called diplotypes to final report on drug recommendations. Here is a link to a recent webcast demonstrating… Read more »
We have been building up to the final release of VarSeq 2.6.0 for a couple of weeks now, but we are excited to announce that 2.6.0 is officially available! VarSeq 2.6.0 is an exciting release as this version of VarSeq features the introduction of VSPGx, offering a complete pharmacogenomic workflow, including data import, variant analysis, and report generation. We have… Read more »
VSPGx is a pharmacogenomics interpretation software based on CPIC recommendations. The field of pharmacogenetics bridges genetics and pharmacology, with the aim of optimizing drug therapies for individual patients based on their unique genetic makeup. It has the potential to revolutionize healthcare by improving drug efficacy, reducing adverse reactions, and advancing the concept of personalized medicine moving away from a one… Read more »
Today, we will be exploring the indispensable role of GenomeBrowse in your VarSeq workflows. This blog aims to guide you through various customization options, including color preferences, filtering techniques, display modifications, numeric value plotting, and additional tips. I hope to enhance your GenomeBrowse experience and enable you to gain valuable insights into your data. Let’s jump in by talking about… Read more »
Thank you to those who attended our recent webcast on Golden Helix’s SNP & Variation Suite (SVS) and its new capability related to Polygenic Risk Scores (PRS). If you were unable to attend, a recording can be found via this link. For common diseases, PRS can provide a predictive value related to the disease risk at an individual level. By… Read more »
As many of you may already know, we just released VarSeq version 2.5.0 this month! We have talked about the two headlining features a bunch, but we have not focused on what else has changed in VarSeq 2.5.0 that might also strike your fancy. For those of you who might just now be tuning into the hype around 2.5.0, it… Read more »
Greetings VarSeq users! VarSeq 2.4.0 has officially been released. VarSeq 2.4.0 is a significant release that focuses on enhancing the VSClinical ACMG workflow by introducing new features and noteworthy improvements. The major highlights of the release are: 1. Welcoming structural variant support to the VSClinical ACMG workflow 2. ACMG workflow automation has been enhanced via the application of evaluation scripts… Read more »
Use the force of evaluation scripts to automate and customize your VSClinical ACMG workflow in VarSeq 2.4.0. VarSeq 2.3.0 came packed with new features! Most notably, VarSeq variant analysis expanded to support the import and annotation of structural variant files, and the AMP cancer workflow in VSClinical gained new functionality with the addition of evaluation scripts which help automate and… Read more »
Discover the innovative clinical evidence search of the VSClinical AMP workflow, where patient biomarkers and tumor types are matched to the most relevant data from top sources like CIViC, DrugBank, and Clinical Trials. One of the core features of the VSClinical AMP interpretation workflow is the ability to search third-party data sources to find clinical evidence that matches the patient’s… Read more »
Revolutionize Your Somatic Variant Analysis with Our Cutting-Edge Template for Annotation and Filtering in VarSeq Golden Helix is excited to share our new Comprehensive Cancer Template for somatic variant annotation and filtering, along with the latest version of our software VarSeq 2.3.0! Our latest VarSeq update was specifically focused on getting up to speed with multiple aspects of somatic variant… Read more »
Discover the latest advancements in cancer genomic profiling with the release of VarSeq 2.3.0 We are very excited to announce the release of VarSeq 2.3.0! This release was one of the largest VarSeq releases yet, as it includes a large refactor to the VSClinical AMP cancer module. A primary motivation for the release was focused on the availability and increased… Read more »
Learn about the latest cancer ontology software updates and how they can enhance your research and work. Cancer Ontology An important feature of the VSClinical AMP workflow is the ability to select the correct tumor type for a given evaluation. The selected tumor type is used to identify relevant clinical evidence, including drug sensitivities, resistances, and previous biomarker interpretations. This… Read more »
Thank you to those who attended our recent webcast by Gabe Rudy, Large Scale PCA Analysis in SVS. For those who could not attend, you can find a link to the recording here. While this webcast discussed methods for principal components analysis (PCA) in SVS, including the new capability for performing principal components analysis on large sample sizes, it also… Read more »
We would like to announce that a new version of SVS has been released! The headlining feature of the SVS 8.9.1 release was new functionality for Large Data Principle Component Analysis. A detailed description of this new feature can be explored in this recent blog post: Finding a Few Principal Components Quickly from Data with Thousands of Samples. However, there… Read more »
I’m sure everyone’s favorite winter activity is getting cozy next to a fire, with a nice cup of tea, and reading the VarSeq instruction manual, right? Of course all VarSeq users have read the manual, but if you have not had the chance to review this captivating read, let me entice all of you to do so by announcing that… Read more »
We are very excited to announce that just last week, we released VarSeq 2.2.4! In the past few months, we have been building the excitement for the 2.2.4 version of VarSeq with several webcasts in which we describe some of the headlining features in detail such as the new support for Gene Panels and Gene Lists, PhoRank Clinical, and Customized… Read more »
As we move towards the end of the year, our FAS Team is excited to announce our short blog series highlighting some of the Customization Features in our up-and-coming VarSeq release! The goal of this blog series is to show examples of how generating a clinical report can be customized to accommodate a wide range of functionality. Our December webcast,… Read more »
With the release of VarSeq 2.2.4 just around the corner, I want to detail some new 2.2.4 features that will enhance somatic variant annotation and fusion analysis within VSClinical. A webcast back in July showed some of these updates in action, so if you are looking for some more content on this topic, I would highly recommend checking out the… Read more »
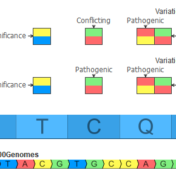
In the September 2021 monthly update to our curated ClinVar track, we made some changes that will result in roughly another 7,000 Likely Pathogenic and Pathogenic variants being available for annotation and use in the ACMG auto-classification system. Consensus Between Labs ClinVar has nearly one million unique variant classification records that are curated into multiple annotation tracks used in VarSeq and VSClinical on a monthly basis. Clinical… Read more »