Thanks to all of you who were able to attend the live webcast introducing the newest genomic analysis tool within VarSeq, VSPGx! For those of you who could not make it to the live presentation and demonstration, I will fill you in on what we covered. If you would like to watch or re-watch the webcast, you can access the… Read more »
In our recent webcast, we unveiled the integration of the Twist Bioscience Exome 2.0 Plus Comprehensive Exome Spike-in capture panel with VS-CNV, marking a significant advancement in genetic diagnostics. By addressing the limitations of standard exome kits that miss vast genomic regions, our enhanced panel introduces ‘backbone’ probes for comprehensive genomic coverage. This innovation enables the detection of CNVs, LOH,… Read more »
Many thanks to those who came to view our most recent webcast, From Panels to Genomes with VarSeq: The Complete Tertiary Platform for Short and Long-Read NGS Data. This was a great opportunity for us to showcase the breadth of our workflows, from short-read to long-read, panels to genomes, singleton analysis to families. In this blog, we would like to… Read more »
Thank you to all our viewers who attended our webcast last week on VarSeq 2.5.0: VSClinical AMP Workflow from the User Perspective. If you did not get a chance to attend but would still like to see the new upgrades to VarSeq 2.5.0, please visit our website here. Overall, this webcast highlighted the versatility of VarSeq, demonstrating both a Tumor-Normal… Read more »
Traditionally genetic tests in cancer have focused on small gene panels that restrict their analysis to a small number of well-studied cancer genes. However, as sequencing costs have decreased, many clinical laboratories have embraced comprehensive genomic profiling tests that rely on whole exome and whole genome next-generation sequencing (NGS) workflows, which can detect millions of high-quality variants for a single… Read more »
With the widespread adoption of next-generation sequencing for clinical and research applications comes the need for guidance and recommended standards and best practices for achieving accuracy and efficacy for each assay pipeline. The support team at Golden Helix has gotten a lot of hands-on experience in assisting users to build their bioinformatics workflows for NGS assays. We recently shared our… Read more »
In our recent webcast, we discussed the exciting new features of VarSeq 2.4.0 and the updated VSClinical interface. The discussion was centered around three main topics: In summary, VarSeq 2.4.0 uniquely supports the analysis of all variant types in the clinical interpretation workflow. By incorporating structural variants, enhancing automation, and empowering users to handle complex data, it offers a comprehensive… Read more »
Explore the cutting-edge capabilities of VarSeq in prenatal genetic screening as we delve into real-life cases, expert analysis, and efficient strategies to quickly assess Whole Exome Sequencing (WES) samples for genetic abnormalities in our recent webinar. Thank you all to those who attended our VarSeq webinar covering prenatal genetic screening! We had a great turnout and loved hearing from our… Read more »
Discover the Thrilling Automation Capabilities of VarSeq Suite for NGS Testing and Learn How We’re Tackling Contemporary Lab Challenges First off, I’d like to thank everyone who joined us for our February webcast on the automation capabilities of the VarSeq software suite through VSPipeline. For those of you who joined us, I’m sure it’s obvious that automation is a topic… Read more »
Our FAS team would like to thank everyone who attended our December 2022 webcast, A User’s Perspective: Somatic Variant Analysis in VarSeq 2.3.0. This webcast allowed three members of our FAS team to give their unique insights concerning the improvements to our new VarSeq 2.3.0 release, which will be recapped here. Starting with template creation, our Technical Field Application Scientist,… Read more »
Thank you to everyone who joined us for our webcast on the upcoming VarSeq features supporting the full spectrum of genomic variation! Traditionally, NGS cancer testing started with small gene panels that looked at a small set of the most common genes to identify small mutations, such as BRAF V600E. However, there are many classes of mutation that cannot be… Read more »
Thank you to those who attended our webcast on the user perspective of our automated AMP guidelines! Furthermore, let me express our appreciation for those particularly engaged users who posed some very thoughtful questions. While we weren’t able to answer all of them live, I hope to shed some light on some pertinent details of somatic analysis here. Let’s start… Read more »
In the era where cloud-based solutions are the default for the modern office, it may not be obvious why many laboratories and testing centers choose to host their data and analysis pipelines on-premises or on self-managed cloud services. In the recent webcast Evaluating Cloud vs On-Premises for NGS Clinical Workflows, I explored the topic of how to make infrastructure decisions… Read more »
Thank you to those who attended our recent webcast by Gabe Rudy, Large Scale PCA Analysis in SVS. For those who could not attend, you can find a link to the recording here. While this webcast discussed methods for principal components analysis (PCA) in SVS, including the new capability for performing principal components analysis on large sample sizes, it also… Read more »
In our previous blog, we covered the highlights of our Advanced Report Customization in VSClinical webcast in the context of germline clinical reports. Now, we bring you the next of the series: somatic clinical reports. In the recent webcast, Advanced Report Customization, we covered a range of somatic-focused clinical reports, demonstrating how easy it is to create AMP guideline-based clinical… Read more »
When it comes to clinical variant analysis for germline variants using ACMG guidelines, we understand that the clinical report is essentially the receipt for your services for the patient. In our recent webcast, Advanced Report Customization in VSClinical, we displayed several examples of report templates to show off the range of possibilities our users have to format their clinical report…. Read more »
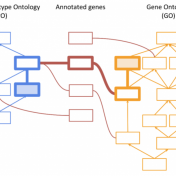
Thank you to those who attended our recent webcast, “PhoRank 2.0: Improved Phenotype-Based Gene Ranking in VarSeq”. For those who could not attend, you can find a link to the recording here. This webcast covered upcoming improvements to the PhoRank phenotype-based gene ranking algorithm based on literature published in the years since the algorithm’s development. The PhoRank Algorithm When performing… Read more »
Thank you to those who attended the recent webcast, “High Precision Exome CNV Detection with VS-CNV”. For those who could not attend but wish to watch, here is a link to the recording. This webcast delved into the complex world of CNV calling for whole-exome samples, which presents unique challenges that require specific considerations and strategies. Over the past several months,… Read more »
Thank you to those who attended the recent webcast, “Exome Analysis with VS-CNV & VSClinical: Updated Strategies & Expanded Capabilities”. For those who could not attend but wish to watch, here is a link to the recording. In this webcast, we covered the capabilities and updates that have been incorporated into VarSeq that enhance whole exome sequencing workflows. The new… Read more »
Thank you to those who attended the recent webcast, “VSWarehouse: Tracking Changing Variant Evidence and Classifications”. For those who could not attend but wish to watch, here is a link to the recording. The webcast covered some general highlights of VSWarehouse value but also presented some specific capabilities covering the ClinVar classification tracker. Golden Helix provides complete solutions to handle… Read more »