While stand-alone SVS is an amazing statistical software package for genomics analysis, adding additional Python scripts to the program can expand SVS’s genomic analysis capabilities. In this post, I’ll take you through the most frequently downloaded Add-On Scripts for SVS and the top five were not what I expected!
Coming in at Number Five is the script Convert Dosages to Genotypes. This script converts allelic dosage values to genotypes based on user-specified thresholds. The dosage data may be in Single or Double-Dosage format. The Single-Dosage format usually has data ranging from 0-2, while the Double-Dosage format has values from 0-1.
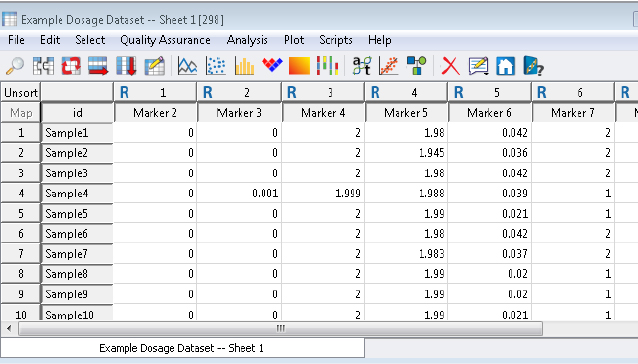 Figure 1. Dosage Data in Single-Dosage Format
Figure 1. Dosage Data in Single-Dosage Format
Number Four is Correct P-values for Multiple Tests. This script takes a column of p-values and outputs several multiple testing corrections including Bonferroni, FDR (Storey 2002), BH FDR (Benjamini-Hochburg 1995) and BY FDR (Benjamini-Yekutieli 2001). The association testing options in SVS give the user the option to output Bonferroni and FDR, but this script includes several additional options. This script can also be run on a set of p-values that were calculated elsewhere and then imported into SVS for continuing analysis.
Number Three on the list is SNP Cluster Plot, which allows the user to create scatter plots based on A and B allele intensities that can be split on SNP genotypes to create tri-colored cluster plots. The script will work for up to 100 SNPs at a time. This script does require the Affymetrix CEL files.
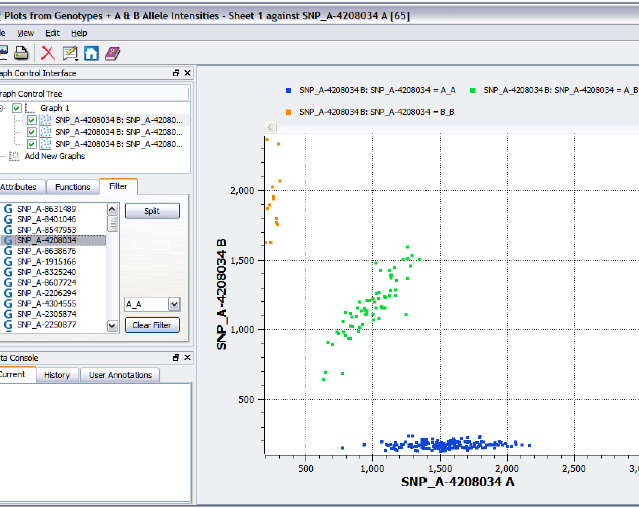 Figure 2. SNP Cluster Plot
Figure 2. SNP Cluster Plot
Coming in at Number Two is a script that calculates Alternate Allele Frequency. This script calculates the percentage of alternate alleles over all samples for each variant. The marker map must contain a reference base field. The resulting spreadsheet has variants as rows and the marker map applied row-wise. The spreadsheet has columns containing the reference count, alternate allele, alternate allele frequency, reference allele count and alternate allele count.
This was a surprising result for me as SVS has this function built in! It’s included in the Statistics by Marker function under the Genotype Menu. When using this option be sure to classify the alleles as Reference/Alternate instead of by Allele Frequency.
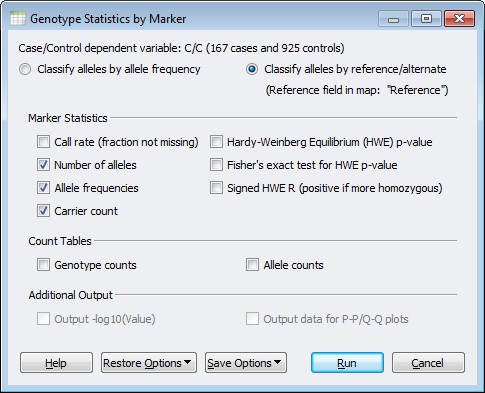 Figure 3. Genotype Statistics by Marker Menu
Figure 3. Genotype Statistics by Marker Menu
And the Number One script downloaded this year from our website – drum roll please! – is Affymetrix B Allele Frequency Calculation! This script uses Affymetrix CEL files as its source and combines quantile normalized SNP A and B probe intensities for each marker into a theta value, then calculates B-Allele Frequencies for each marker.
Golden Helix has many other scripts to facilitate your genomic analysis in SVS; click here to go to our Add-On Script Page and browse the selection.
If you have any questions about any of our add-on scripts please email [email protected] and we’d be happy to assist you!
Great overview. Just wondering what the position was of my script (https://www.goldenhelix.com/SNP_Variation/scripts/pages/ActivateATCGSNPsToFlipStrand.html). Do you know? 🙂
Hi Sander,
Your script is currently the 23rd most used!