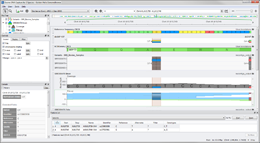
The above screenshot shows the exomes of three species (Bison bison, Bos indicus, Bos taurus) aligned to the Bos taurus UMD 3.1 reference sequence.
In our recent webcast, Advancing Agrigenomic Discoveries with Sequencing and GWAS Research, Greta Linse Peterson featured bovine data which she download from the NCBI website. The data was downloaded in SRA format and in order to analyze the data in SVS, the files had to be converted to BAMs and then merged into a single VCF file. Since many of you are accustomed to wrangling your data on a regular basis (or maybe you leave the wrangling to someone else), we thought we would share the secondary analysis steps we used when preparing the data. Our goal was to run the data through a common, “plain vanilla” pipeline, so that we were not relying on any special features during our downstream analysis. As such, we chose to use a combination of BWA and GATK; common tools that are often used in conjunction with each other.
Step 1: To Start – SRA to FASTQ
As previously mentioned, the files came from the NCBI website in SRA format. To start the process, the SRA files had to be converted to FASTQ files. We used the SRA Toolkit “fastq-dump” command for the conversion since it was provided on the NCBI website.
Step 2: Alignment – FASTQ to BAM
For alignment, we used the Burrows-Wheeler Alignment Tool (BWA). BWA is a widely used and accepted algorithm for alignment, and it strikes a nice balance of speed and accuracy. BWA is about 10-20x faster than MAQ, which used to be a standard alignment algorithm in the bioinformatics toolkit.
In order to align the reads to a reference, an index is necessary – it allows the alignment algorithm to efficiently find all the places where the read may align. To accomplish this, the FASTA files (also downloaded from the NCBI website) were combined and the index was created using BWA. In order to create the index, BWA requires a Burrows-Wheeler Transform of the reference sequence to be computed, which then enables the alignment algorithm to be quite quick.
After combining and indexing the FASTA files, BWA was used again to align the samples with the bovine reference sequence, which consisted of two sub-steps. First, the reads are aligned using suffix array (SA) coordinates using the “aln” command. Then the SA coordinates are converted into genomic coordinates using either the “samse” or “sampe” sub-commands. This two-step process is useful since reads can be either single-end or paired-end. In both cases, we can use our previously created index to generate SA coordinates for each read. However, when converting SA coordinates to genomic coordinates, we can use the fact that paired-end reads should be a known distance apart to help increase the accuracy of the alignments. Since the bovine reads were single-end, we used the single-end algorithm for the conversion to genomic coordinates.
Because GATK requires a read group in the BAM header, we set it as the sample name during the creation of the SAM files. Alternatively, Picard provides a utility that can be used later to edit this element of the SAM header.
Finally, we used the “view” command in SAMtools to convert the SAM files into BAM files. BAM files contain exactly the same information as SAM files, but use a binary file format rather than text format to reduce the file size. To index these BAM files, SAMtools requires that they be sorted in order of coordinates. Since the BAM files output by the aligner are in the order that the reads were processed, we used the sort utility provided by the SAMtools package just prior to indexing the BAMs.
Step 3: Realignment – BAM to better BAM
After the alignment was complete, the GATK IndelRealigner was used to perform a local realignment on the BAM files to minimize mismatches. Mismatches occur when a base in your read does not match the reference. It could be a sequencing error, misalignment, or it could be a real mutation. Since indels can shift the reads by a base or two, they can throw off the alignment of the entire read thereby causing one or more mismatches. These mismatched reads can decrease the accuracy of our final variant calls.
Step 4: Variant Calling – BAM to VCF
Finally after downloading, aligning, realigning, sorting, and indexing, we have BAM files that are a good representation of the sequenced samples and are ready to call variants using the GATK UnifiedGenotyper.
One additional input that GATK requires is a sequence dictionary; this file can be created using Picard.
Optionally, GATK allows the user to use the dbSNP catalog during variant calling. This will provide you with rsIDS in your final VCF. However, in order to use this feature you must make sure that the contig names are consistent between the reference and the dbSNP catalog; by default they are not. We fixed this using the command line utility Sed. Note that the ordering must be consistent between the two sources as well.
Since wrangling command line tools isn’t everyone’s idea of an exciting Friday night, we could have skipped using a dbSNP catalog during variant calling and instead added rsIDS to our variants using SVS.
Our end goal was to obtain a single VCF file, so we passed in the previously created BAM files for each sample as inputs to a single run of GATK’s UnifiedGenotyper. Ninety minutes later, after allowing our 8 cores to churn through the data, we have our single, multi-sample VCF file!
As any bioinformatician knows, a large part of the job is simply getting the output of one tool into the format required by another tool. And while this analysis can be tedious and time consuming, it’s well worth the effort when you get exactly what you need. With a little practice these steps become second nature, and you can start to build out scripts to automate and parallelize much of the process. Good luck!