Use the force of evaluation scripts to automate and customize your VSClinical ACMG workflow in VarSeq 2.4.0.
VarSeq 2.3.0 came packed with new features! Most notably, VarSeq variant analysis expanded to support the import and annotation of structural variant files, and the AMP cancer workflow in VSClinical gained new functionality with the addition of evaluation scripts which help automate and customize the creation of evaluations. Now, I know what you are all thinking; I wish these amazing features could be available for germline analysis within VSClinical ACMG workflows… with the upcoming VarSeq 2.4.0 release, you can consider your wish granted!
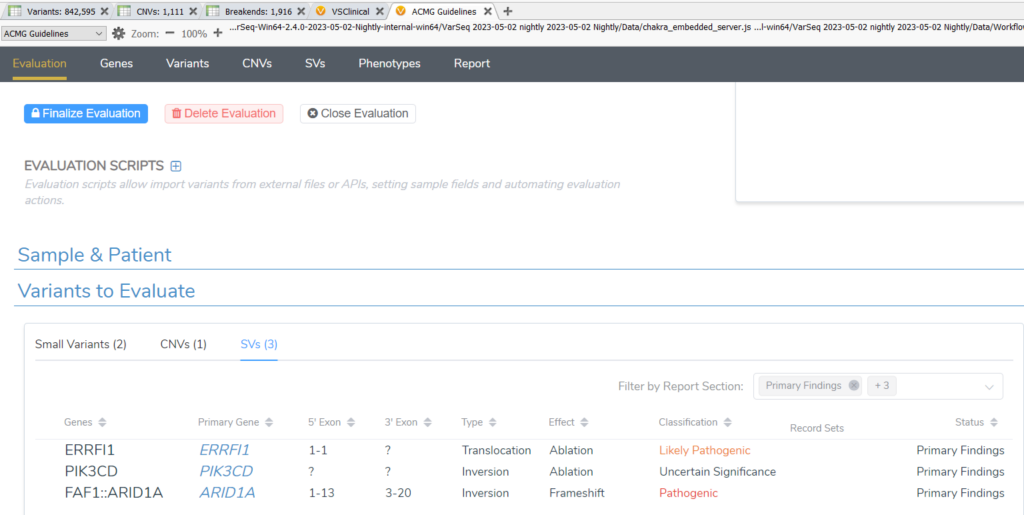
Let’s start by introducing the new structural variant support for the ACMG workflow. VarSeq 2.3.0 enabled the import of structural variant files to their own variant table, which can be annotated with gene details, effect predictions, gene panels, and more. In VarSeq 2.4.0, those annotated structural variants can be imported directly into VSClinical ACMG or manually added to evaluations. A new assessment catalog dedicated to structural variants has been added so classifications and interpretations can be captured for SVs in addition to existing catalogs for small nucleotide variants and CNVs. You will likely notice that the evaluation tab has experienced a bit of a face-lift, but the design is meant to facilitate analyzing the three types of variants seamlessly and accommodate the evaluation scripts.
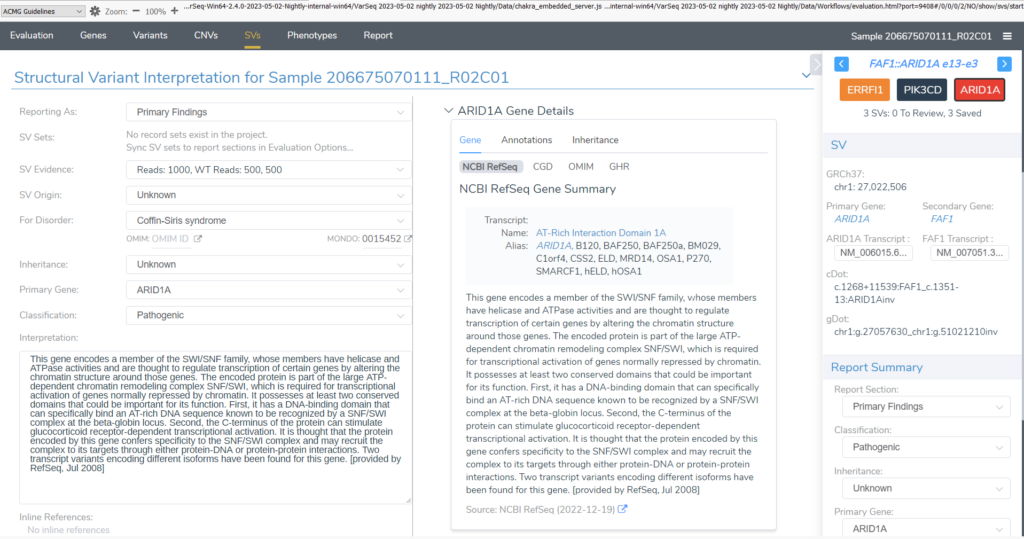
Structural variant analysis in VSClinical ACMG is very similar to analyzing variants and CNVs. In the SV tab, you can set report status, classification, associated disorder, and interpretation for SVs just like can be done for variants and CNVs. To bring the analysis full circle, the report templates have each been updated to support reporting structural variations.
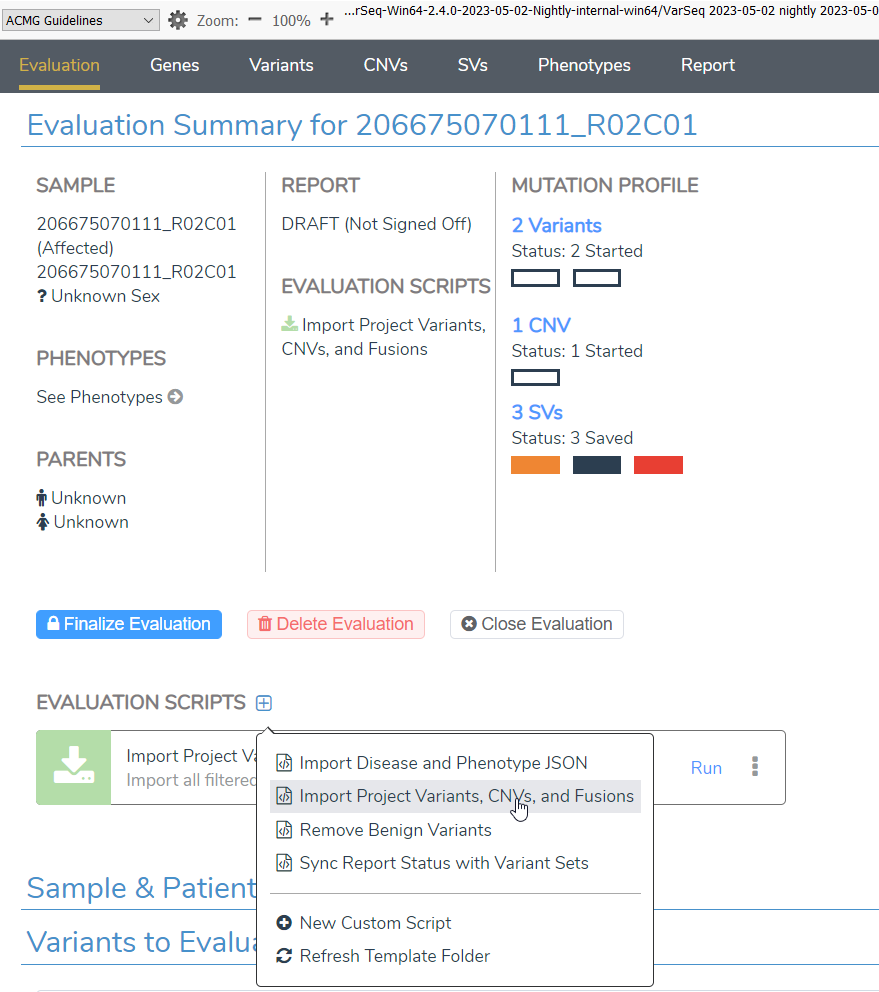
Now that we are all stoked on structural variants, I want to sweeten the pot by talking about evaluation scripts! Despite their intimidating name, these nifty “scripts” are quite user-friendly and enable a lot of automation functionality, saving you time and effort. These evaluation scripts were introduced to the VSClinical AMP workflow in VarSeq 2.3.0, and VarSeq 2.4.0 will bring them to ACMG! Below is a list of the evaluation scripts that we will have available. However, the best part about the evaluation scripts is that you can modify the scripts or even make up all new scripts of your own that can integrate with VSClinical.
- Import Project Variants, CNVs, and Fusions script add the filtered result of the variants, CNV, and breakend tables to a new evaluation.
- Sync Report Status with Variant Sets script enables record sets from variant tables to be used to define the report status of the variants that have been added to the evaluation (i.e., record sets mapped to “Primary Findings” will get a report status of “Biomarker”, record sets mapped to “Secondary Findings” will get a report status of “Secondary Germline”, and record sets mapped to “Uncertain Significance” will get a report status as “VUS”).
- The Import Disease and Phenotype JSON script can import sample disease and phenotype data from JSON file input. To use this script, the imported files must be formatted based on the phenopackets schema. An example script/schema can be found within the script’s folder. Click on the vertical 3 dot menu for the script and click Open Location to access the script’s folder.
- The Remove Benign Variants script will remove any variants, CNV, or SV that are classified as benign after they have been added to an evaluation.
You can add evaluation scripts to individual evaluations or incorporate them into VSPipeline workflows to take you from variant import through VSClinical evaluation and to generating a clinical report.
Now I make it seem like these are the only features that we can expect to see in VarSeq 2.4.0, but that is not the case. We have much more to look forward to in VarSeq 2.4.0, and we have a webcast scheduled for May 17th that will introduce VarSeq 2.4.0 in all its glory. Until then, May the 4th be with you all!