In our previous blog, we covered the highlights of our Advanced Report Customization in VSClinical webcast in the context of germline clinical reports. Now, we bring you the next of the series: somatic clinical reports. In the recent webcast, Advanced Report Customization, we covered a range of somatic-focused clinical reports, demonstrating how easy it is to create AMP guideline-based clinical reports and to bring in custom fields into the final clinical report. Below, we will review a standard somatic clinical report and an example of report customization.
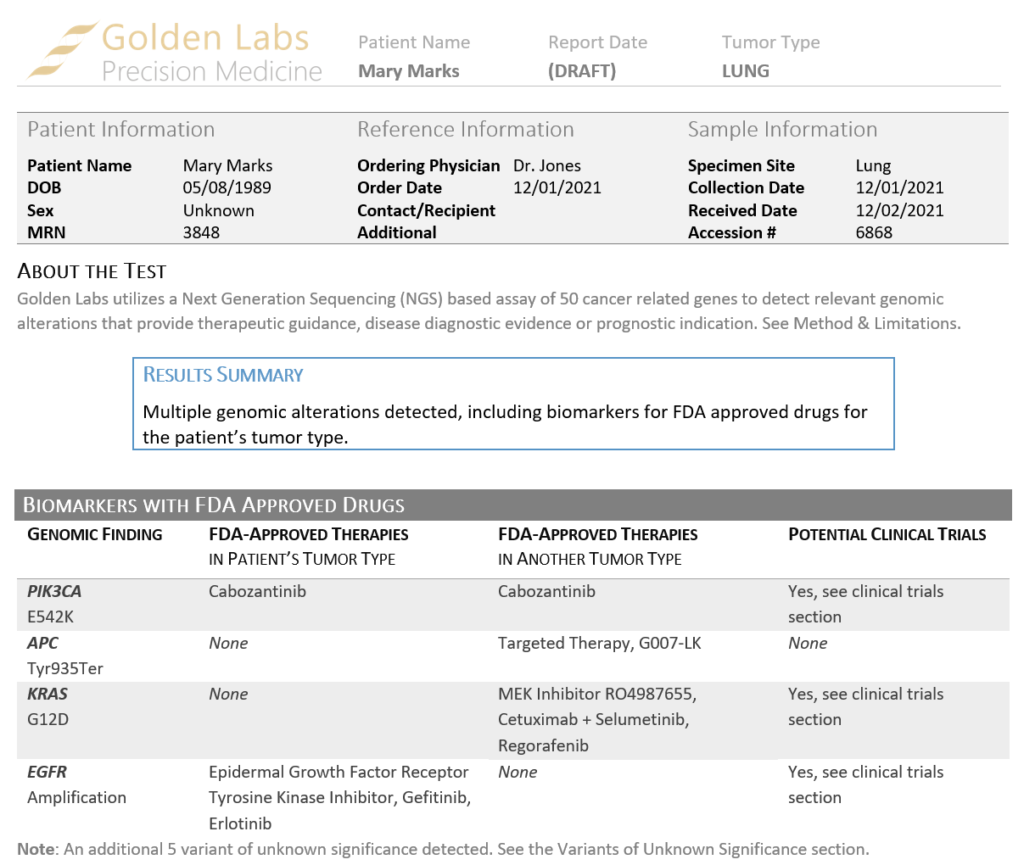
There are two somatic templates shipped with VSClinical AMP. The simpler Cancer Report template is focused on biomarker and drug information, while the Cancer Drugs and Trials template allows users to also include clinical trial information. In the recent webcast, we reviewed our Cancer Drugs and Trials template, and several styles of custom clinical reports based on biomarkers identified in a Lung Carcinoma context, including a single nucleotide variant in the gene PIK3CA, which will be the focus of this article. Similar to the ACMG-based report templates reviewed in our previous blog, our somatic clinical reports have several key fields that can be brought in from a sample manifest, including patient demographic information, physician information and specimen details. This layout also displays a summary of the results and a list of significant biomarkers with available treatment options (Figure 1). These fields and the company logo can all be easily customized to suit your needs.
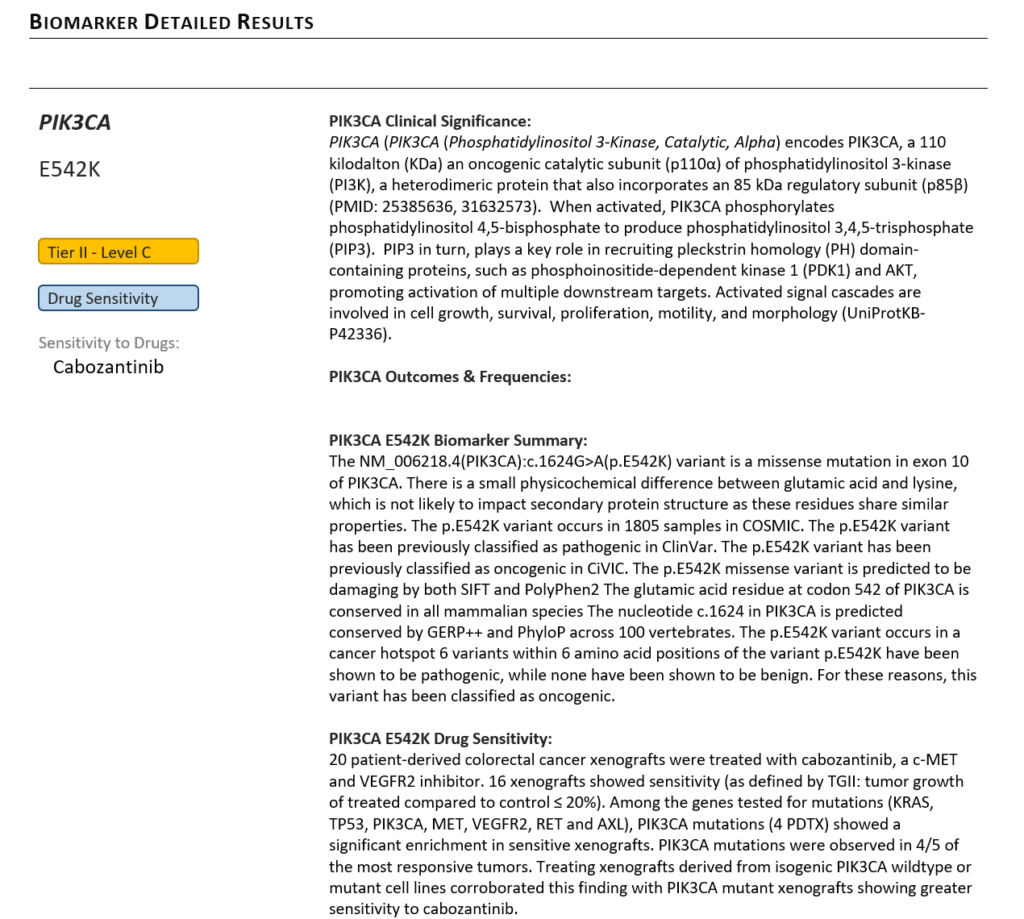
Both of our AMP-based standard clinical report templates have sections detailing the information brought in about relevant biomarkers and assessing available drugs. VSClinical automates the population of all relevant biomarker evidence into the final clinical report, including AMP Tier level details, clinical significance, and drug sensitivity assessments (Figure 2). Golden Helix provides the predefined clinical assessments of biomarkers via our CancerKB catalog which is kept up to date by our team of curators. For more information on using CancerKB to accelerate NGS analysis, you can watch this recent webcast.
Additionally, when using the Cancer Drugs and Trials Template, VSClinical automatically renders sections for the drugs and clinical trials matching relevant biomarkers. This section includes detailed information on the clinical study, trial site, contact information and patient eligibility (Figure 3). Users can format their clinical report to include as much of this information as is relevant for their reporting needs. These examples are just one layout that can be used to report on the variants that come through your VSClinical evaluation.
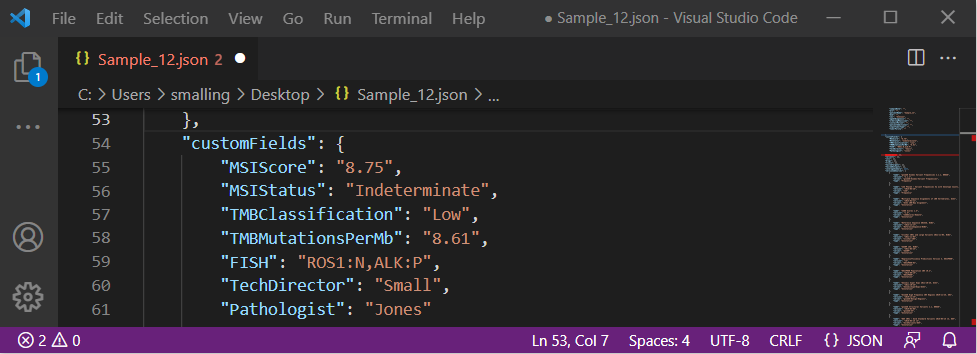
We also recognize that the ability to bring in custom fields unique to somatic testing is often desired in the context of AMP guideline-based clinical reports. Some of these fields include microsatellite instability (MSI) status and tumor mutation burden (TMB) classification. These fields can now be brought in from a custom sample manifest, viewed in the samples table and json file (Figure 4), and ultimately brought into the final clinical report.
In order to bring in custom fields into the clinical report, users can employ a few simple scripts in their clinical report template. There are options to include, for example, signatures of members of your clinical team (Figure 5), and additional non-NGS companion diagnostics relevant to a somatic workflow such as PDL1 immunohistochemisty (IHC) results.
In our next blog in the series, we will explore additional customization options that are accessible to users for both VSClinical ACMG and AMP-based clinical reports. You can read more on our VSClinical ACMG or AMP workflows and report customization in our other blogs and webcasts. For help with any advanced customization of your clinical reports please reach out to us at [email protected].