It’s the big day, and you’ve spent hours preparing for the big old-fashioned family Thanksgiving. Keeping in mind that your Cousin Eddie likes to show up unannounced and eat you out of house and home, you have remembered our food allergy allele workflow from last year, and prepared a special menu. Right on time, Cousin Eddie arrives, family in tow. Unlike last year, now he is complaining about a bad case of Gastroesophageal reflux, better known as acid reflux. As his son, Rocky, is also complaining about GERD, you wonder if this is a genetic condition caused by far more than Eddie’s sub-optimal dietary habits. Breaking out your new and updated VarSeq 2.6.2, it’s time to return to Eddie’s genetic sequencing.
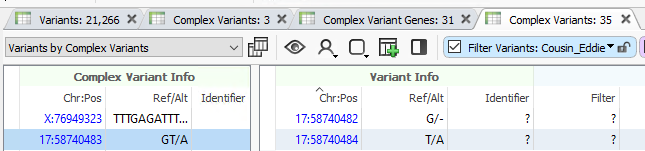
In the last year since we’ve taken a look at Eddie’s genetic screen, VarSeq has undergone a number of updates. These include, and are not limited to, the ability to leverage phasing to look for collapsed phased variants, and a new Complex Variant table. Previously, VarSeq would have taken a Complex Variant and split it into allelic primatives. For example, a variant of GT/A would have been split into respective G/- and T/A. This may have been more convenient for overall filtering, but there would not have been a good way to look at the overall impact of the G/- and T/A separately and together. In VarSeq 2.6.2, we now have the ability to view and analyze the complex positions together and separately to look for the combined impact (Figure 1). Keeping the Complex Variants table in mind, we can now upload Cousin Eddie’s VCF and look for Complex Variants that we may have previously missed.
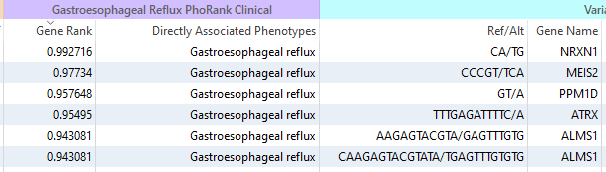
The exciting news for us, not so much for Cousin Eddie, is that we see a number of Complex Variants now retained in our VarSeq workflow. The goal, like with any NGS workflow, is to get down to the clinically relevant variants. So after applying a set of quality filters, I am now able to apply my HPO Terms. Using PhoRank, I am searching for variants in Genes associated with Gastroesophageal reflux, or HP:0002020 (Figure 2).
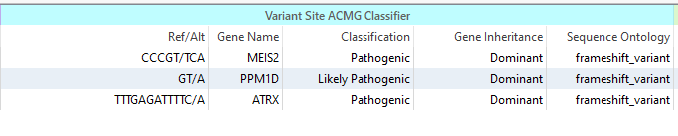
Our next goal will be to sort for the Pathogenic or Likely Pathogenic Variants, which can be done with our ACMG Classifier. Here we see three different variants that meet that criteria (Figure 3).
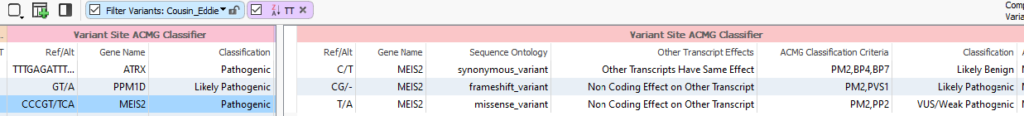
While all three of those Pathogenic Complex Variants are interesting, the MEIS2 variant really takes the cake (or in this situation, the pumpkin pie!). The conserved complex variant has a Pathogenic rating, while when the variant is split into three individual representations, we see a Likely Benign variant, a Likely Pathogenic Variant, and a VUS/Likely Pathogenic variant (Figure 4). Therefore, it makes the most sense to import the MEIS2 variant into VSClinical for further analysis in its Complex State.
Our new tools have really come in handy while looking for Gastroesophageal reflux associated Complex Variants for our beloved Cousin Eddie. Now with all this new information in hand, will he be more cautious when it comes to his holiday dietary habits? You’ll have to wait for the next blog to find out!
From all of us at Golden Helix, have a great Thanksgiving weekend!