Recently, we have written a couple of blogs that were talking about the use of Evaluation Scripts in VSClinical workflows, in particular for the AMP workflow. Evaluation scripts were first introduced to VSClinical AMP in VarSeq 2.3.0 and VarSeq 2.4.0 welcomed evaluation scripts to the ACMG Workflow. Evaluation scripts are a nifty way to customize your workflow and say import specific data such as phenotype json or TSO 500 files or automate optimizations to your workflow like to remove all benign variants from an evaluation. Evaluation scripts are also helpful for keeping VSClinical flexible as we can continually add new evaluation scripts or you can create your very own scripts with our help or all on your own!
Since evaluation scripts are a new feature to VSClinical ACMG, I wanted to talk a little bit about each one and how you might be able to use them to either optimize your current workflow or help you build a new workflow if you are just starting out.
VSClinical ACMG Evaluation Scripts:
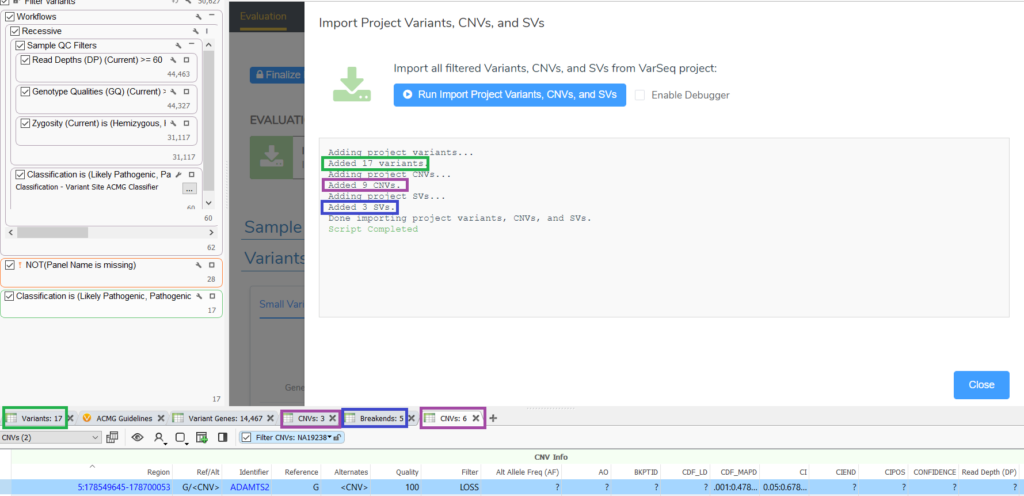
- Import Project Variants, CNVs, and Fusions script adds the filtered result of the variants, CNV, and breakend tables to a new evaluation.
- Running this script allows you to add the filtered results for each variant type (small variants, CNVs, and breakends) from VarSeq tables in only one or 2 clicks. This script is also useful if you have filtered variants from multiple small variants, CNV, or breakend tables as it will add the results from all of the tables (Figure 1). Without this script, you would first click on the “Small Variants” tab of the Variants to Evaluate table, click “Add Variants From Project,” click “Prepare to Add,” choose the “Score All Recommended Criteria” option, then click “Add Variants,” then wait for the variants to populate the Variants to Evaluate table. Then, repeat these steps for CNVs and breakends.
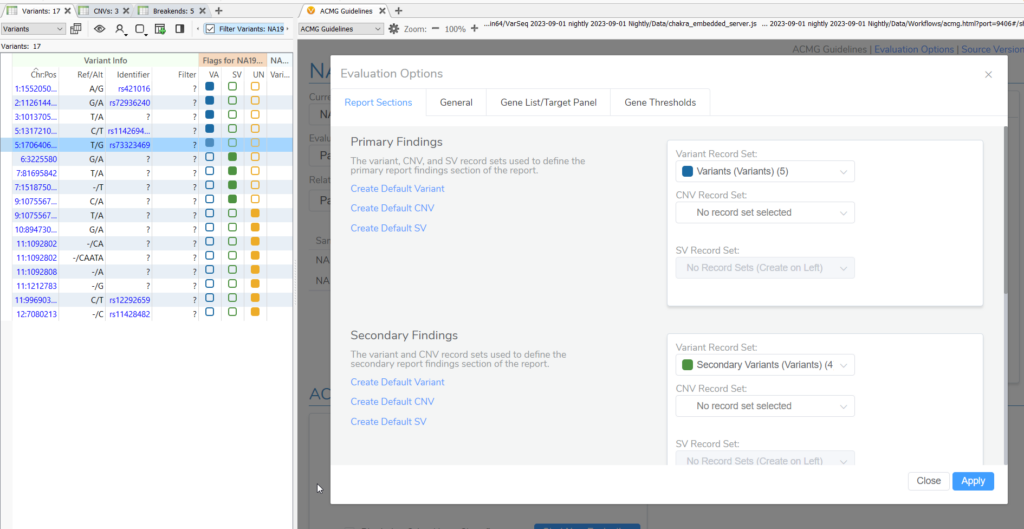
2. Sync Report Status with Variant Sets script enables record sets from variant tables to be used to define the report status of the variants that have been added to the evaluation.
- Before running this script, either create or map existing variant record sets to “Primary Findings,” “Secondary Findings,” or “Uncertain Significance” in the Evaluation options. The record set that is mapped to each Primary, Secondary, or Uncertain significance will be included in that report section (Figure 2). After the variants to the evaluation, then run this script to sync the report statuses. Without this script, you would have to manually set the report status for each variant individually as by default variants are set to “Do not report”.
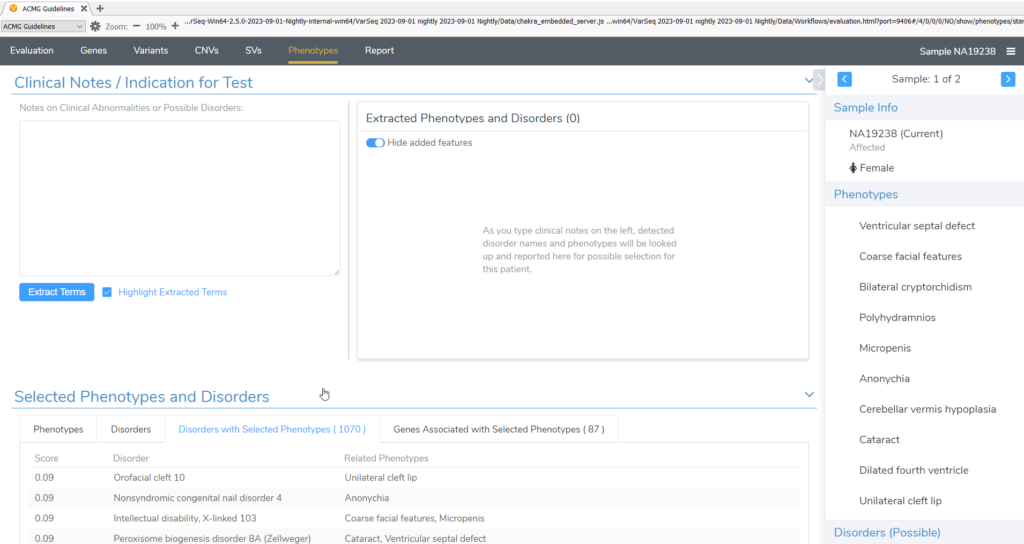
3. The Import Disease and Phenotype JSON script can import sample disease and phenotype data from JSON file input.
- To use this script, the imported files must be formatted based on the phenopackets schema. An example script/schema can be found in within the script’s folder. Click on the vertical 3 dot menu for the script and click Open Location to access the script’s folder. After running this script, the entered phenotypes and disorders will populate in the phenotypes tab (Figure 3) and in the sample clinical report.
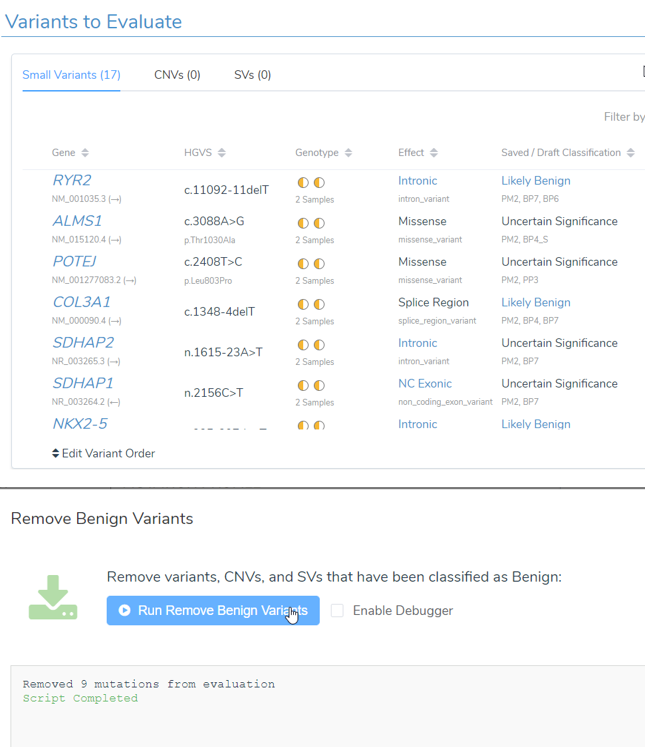
4. The Remove Benign Variants script will remove any variants, CNVs, or SVs that are classified as benign after they have been added to an evaluation.
- This script was added by popular demand as many users wanted to remove any variants that have been classified as benign from the evaluation in bulk! This script is useful if you want to add many variants, CNVs, and SVs automatically to a VSClinical evaluation and you want to report VUS and Pathogenic variants only based on their autoclassification or previous internal classification.
These are the evaluation scripts that are currently available for use within VSClinical ACMG. Keep your eyes peeled for a new evaluation script that will be available in VarSeq 2.5.0 that will allow you to import a database of previously evaluated variants to a VSClinical assessment catalog in one swoop!