The potential of genetic testing to impact a patient’s life has been greatly accelerated by the sharing of variant interpretations done by clinical labs in public repositories such as ClinVar. This is not an inevitable outcome, but the persistent work and advocacy of people like Dr. Heidi Rehm and organizations like ClinGen. We recently participated in a survey and vetting process that ClinGen put together for the clinical lab community in which commercial software vendors support the data sharing standards they define. After vetting and reference checking participating vendors, ClinGen has now published its Genomic Analysis Software Platforms list.
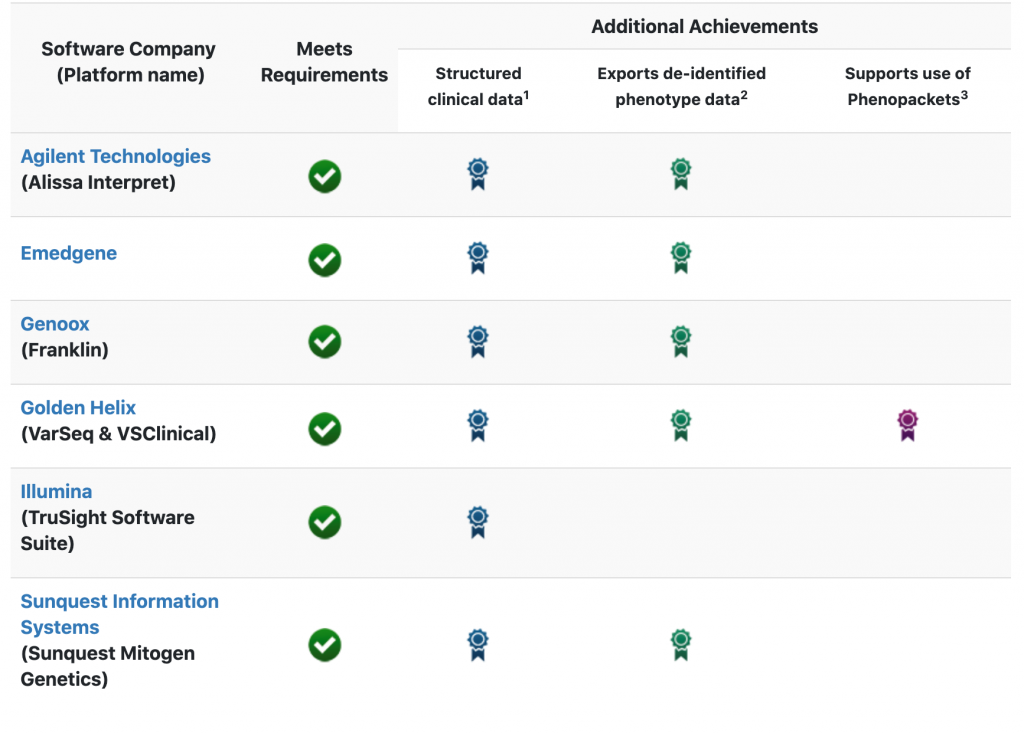
The software companies supporting ClinGen’s data-sharing requirements as of August 2020
Golden Helix has worked closely with the ClinVar team over the years to ensure that our monthly releases of annotations, with the latest ClinVar variants and CNVs, contain the most complete and useful information for our product. Our clinical interpretation workflows in VSClinical pull in ClinVar classifications, clinical assessments, and gene-level counts into the recommendation engine and interactive criteria supporting plots and tables. We are always impressed with how much is added to ClinVar on a monthly basis. Beyond that, the data is constantly evolving, with improved genomic representations, cleaned up duplications, and updated classifications.
The minimal requirements for meeting the data-sharing software vendor list included the ability to submit the work done in the clinical analysis platform to ClinVar. Our VSClinical assessments are always saved into a shared catalog, and we have recently added the support for exporting selected entries into the specialized format needed for ClinVar submission. If you would like training on how to perform this export, please contact our support team!
Beyond the minimal requirements, there were three additional achievements possible for software vendors. We are pleased to say that we are currently the only software vendor on this list to meet the requirements for all three of these additional achievements! These achievements were determined by the software platform supporting the following capabilities:
- Structured clinical data: The ability to import relevant clinical information into the analysis using structured data such as HPO terms, MONDO IDs, etc.
- Exporting de-identified phenotype data: The ability to export the results of the interpretation and evaluated phenotypes with relevant structured terms (from HPO, MONDO, etc) in a de-identified way.
- Supports use of phenopackets: Export not only the variants but the case-level data evaluated for the patient using the structured data format defined by Phenopacket schema.
We want to thank ClinGen and Dr. Rehm’s continued work to push forward the state of data sharing in the clinical genomic world. We also want to congratulate each and every one of the vendors that participated in this process and made the supported platform list. Of course, it is the clinical labs themselves that generously contribute their work and patient data to these repositories that allow for rare variants and rare diseases to in aggregate have actionable and meaningful clinical outcomes for genetic tests. As NGS testing continues to grow and expands to include new classes of variants such as CNV, we expect these requirements and data sharing guidelines to evolve. We look forward to continuing to work with ClinGen and ClinVar to provide our clinical lab users with the best possible solution for variant analysis as well as the long-term storage and sharing of the resulting interpretations.