We have now reached the final blog of the NGS-Solutions for Clinical Variant Analysis series. Part I of this series explored the capture of variant classifications in the VSClinical environment when following the ACMG and AMP guidelines. Part II was similar in content but for the capture of clinically relevant copy number variants as well as using a CNV catalog to easily isolate and eliminate false positive events due to secondary analysis artifacts. So far, we’ve discussed the more detail-oriented steps behind the creation of the catalogs and the kind of detail stored in the catalog itself. Now we are going to elaborate a bit more on sharing this content amongst users performing the variant analysis.
It is a challenge to thoroughly identify and capture all levels of relevant evidence for the determination of variant impact and classification. Once captured, sharing this content internally with all other technicians is crucial for entire workplace efficiency and maintaining accuracy with interpretations. Over time, whole genome scale data is likely to become the norm, and handling the millions of variants per sample is going to require every tool available for optimizing workflows. One important tool Golden Helix provides is VSWarehouse as a genomic repository. With VSWarehouse, users can store all variants seen across their entire cohort of samples which serves as a frequency filter to prioritize rare variants seen among your own pipeline.
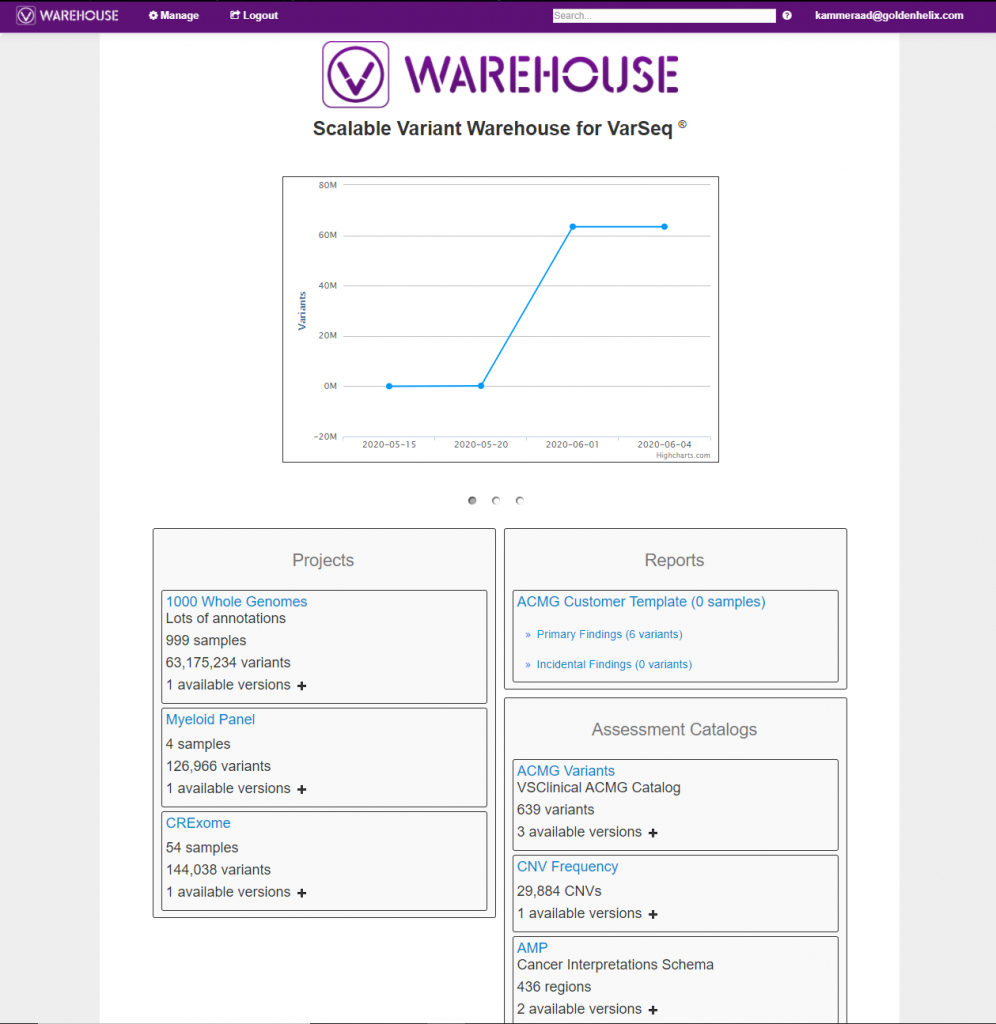
Content to be stored in VSWarehouse includes project data for all variants, assessment catalogs for captured clinical variants, clinical reports, and sample data, which can also be extracted through a hospital LIMS system. Once stored, all this captured data can be managed for permissions at an individual field level, rapidly queried for possible export of interesting variants, and audited with clinical databases like ClinVar to easily assess if variant classifications change over time for your stored variants. Figure 1 illustrates the primary VSWarehouse browser users can access to manage/search through all their stored content. Also, notice the scale of variants stored in the example VSWarehouse instance is over 60 million while still managing speedy searching via the browser
Figure 1. Browser designed to explore the stored genomic data in VSWarehouse.
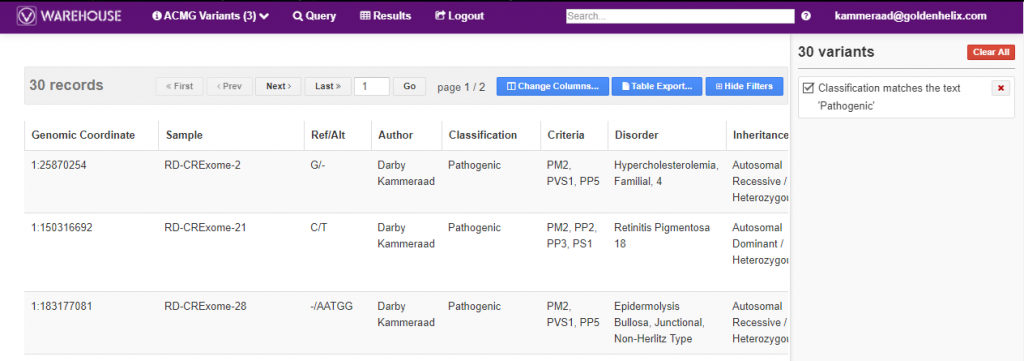
Variant queries can be made for any stored content, which, of course, includes the assessment catalogs. You can see in Figure 2 the results of querying for pathogenic variants stored in the ACMG catalog across 639 captured variants (see Figure 1 above). This browser will also list all the fields users would wish to browse or even export. Exporting the data in perhaps an excel format or even VCF makes for simplicity if wanting to audit the captured variant interpretations/classifications in VarSeq.
Figure 2. Searching through the VSWarehouse ACMG assessment catalog to review ‘pathogenic’ variants.

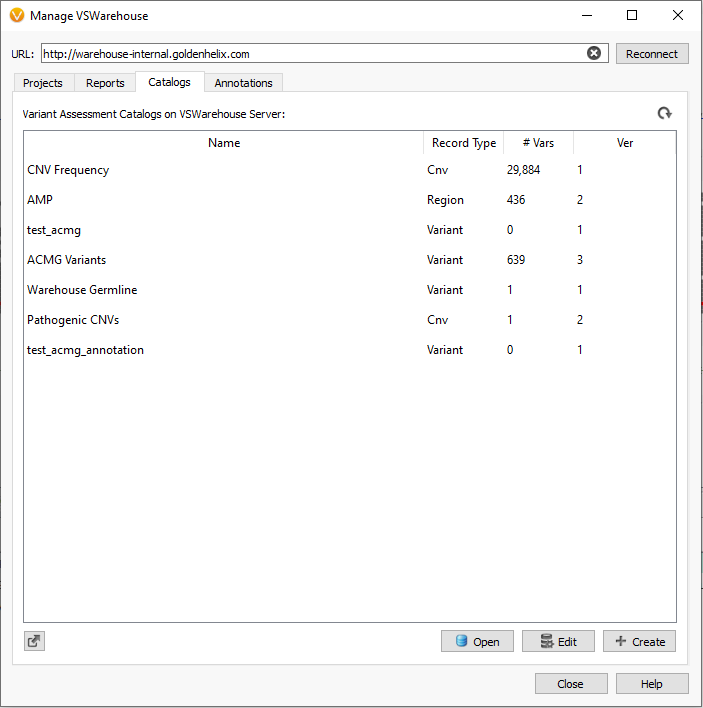
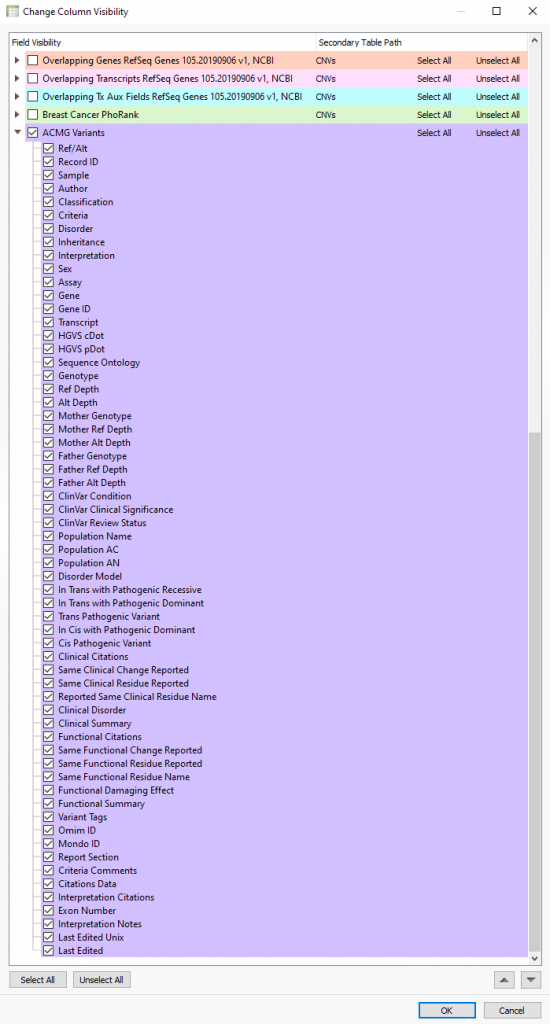
Moreover, the data stored in VSWarehouse is also leveraged in VarSeq as well. This includes using project data like the 60 million variants mentioned previously being leveraged as a filterable field to eliminate variants commonly seen among your cohorts. This is, of course, also relevant in using allele frequencies to eliminate secondary artifacts, as was the case for the CNV frequency catalog example given in Part II of this blog series. From the VarSeq software, users can open the VSWarehouse terminal to access and utilize all the store data (Figure 3). Not only can the contained data be used for filtering, annotating, and visualization, but this becomes the shared content across all users accessing VSWarehouse. For example, each user running a new sample through VSClinical would process the variants, capture the interpretation/classification in the assessment catalog (Figure 4) and push the new variant and data up to the VSWarehouse catalog. Then the next user accessing the VSWarehouse catalog would be able to leverage that recently added variant if they see it in another sample or if assessing new variants nearby in the same gene/exon. Figure 5 illustrates the fields captured in the catalog on a per-sample basis so to review not only all variant level detail, but also quickly assess which samples the variants were found.
Figure 3. VSWarehouse terminal in VarSeq to create and upload projects, clinical reports, and catalogs.
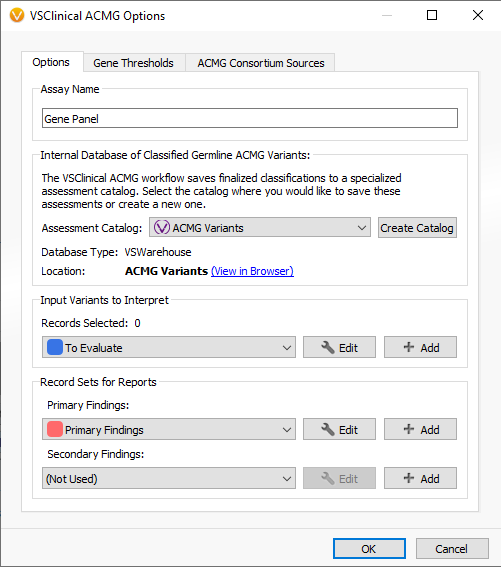
Figure 4. Accessing the VSWarehouse based assessment catalog to centralize the capture of variant classification/interpretation.
Figure 5. Default field captured in the ACMG based assessment catalog presented from the VarSeq variant table.
These catalogs offer huge value in the stored, management, and efficiency of NGS-based variant analysis and VSWarehouse amplifies this value even more by centralizing the data for query and review across all users in a lab or across labs. This blog is merely meant to be a snapshot of demonstrating this value. It goes without saying that for our future users to really understand the value, the software needs to be tested and training on workflow strategies needs to be scheduled. Golden Helix is well known for our high-quality support experience and our Field Application Scientist are always ready to assist you with a trial of the software. Please do not hesitate to reach out to [email protected] if you are interested in learning more.