There is nothing cooler than having something arrive that you have been excitedly waiting for: last week I got an email notification that my 23andMe exome results were ready.
Actually, I got 3 emails that my exome results were ready.
You see, I lucked out.
It all began two years ago on DNA day when Hacker News reported that 23andMe was running a special deal on their personal SNP-array genotyping and interpretation service. Looking across the room at my 7 month pregnant wife, I smiled and pulled out my credit card.
I then proceeded to enter in its numbers – 3 times.
Thankfully 23andMe allowed for returning your spit-in-a-tube DNA up to 6 months from the purchase of your order. Given one of my spit providers was my yet-to-be-born son, this was a very fortuitous policy.
Minus the frustrations of getting a newborn to provide what seems like a million little droplets of spit, the 23andMe customer experience turned out to be really quite entertaining and useful.
For example, based on the roughly 1 million SNPs, dispersed across the genome, 23andMe can predict fun traits like earwax type and the ability to taste bitter food. They can provide useful pharmacogenomic assessments like your sensitivity to Warfarin and how fast you metabolize caffeine. Finally, they can predict your lifetime risk of contracting common genetically-linked diseases such Coronary Heart Disease, Type 2 Diabetes, and certain types of cancers.
But as I talked about in my recent post, the research that 23andMe’s predictions are based on have serious limitations as to how much of the real genetic risk for these complex diseases it can account for with the common SNP-based genotyping it provides with its service.
There will probably be many ways in which further research and sequencing techniques will account for this missing genetic risk, but a promising research direction is examining rare variants in and around the protein-coding genes (the exome) that have the potential to directly influence biological function.
Sequencing human exomes has recently become affordable with Next Generation Sequencing. And more importantly, this technology has shown in many cases to be an effective tool to diagnose rare, serious, and sometimes deadly genetic disorders.
By sticking to the common SNPs genotyped on a custom microarray, 23andMe has been keeping to the previously researched and well-annotated pieces of genomic information.
In contrast, exomes have the potential to uncover not only rare and clinically relevant variants, but in nearly every case are likely to uncover damaging variants of unknown significance.
Stage Left: Your Exome
It is not something I saw through an advertisement or other direct promotion by 23andMe, but at some conference or talk I heard from within the genomic community: 23andMe will sequence your exome for $1,000!
Immediately, I thought: “That’s cool, but my exome is probably pretty boring… What good would an exome be without some interesting angle to investigate?”
Then I realized, there are two exomes I would be very excited to analyze: my wife’s and my son’s.
You see, my wife was diagnosed as a teenager with a relatively rare autoimmune disease. While the symptoms and progression of her disease are managed with the amazing advances of biologic-based therapies like Enbrel, the genetic architecture of immune disorders are complex and suspected to be highly influenced by rare and private mutations.
And, like many fathers, I have an insatiable curiosity about everything that has to do with my son.
What could be more indulgent of that curiosity than being able to browse his DNA? I imagined, at times, that by watching lists of variants and genes scrolling by, I’d get insights into the fascinating, complex creature I watch with loving obsession each day.
So then the idea came to me, and I couldn’t let it go.
I have a trio I could analyze!
Third Sample Is the Charm
There are good reasons why clinical researchers that diagnose rare Mendelian diseases almost always ensure they can sequence not only the affected child but the father and mother as well (and preferably have the option to include living grandparents as needed).
A trio (a father, mother, and child sample set) gives you a lot of unique analytic capabilities. For example, you can use the fact that all variants in the child should either be inherited from a variant in the mother or the father to filter out spurious or low-quality variant sites in the child.
Well, unless the variant is actually occurred de Novo. But given the natural mutation rate of humans from generation to generation, we only expect to see, on average, about 1 de Novo mutation in our exomes per generation.
A trio can also be used to analyze the inheritance of variants, such as when two carrier parents pass on their risk allele to a child.
As you can imagine, this opens the door for new and exciting things to discover and speculate about.
Given the 23andMe exome pilot project by default limited you to one exome per account, I was very excited when they agreed to sequence my whole trio. (When talking to Brian from 23andMe at the TCGC conference recently, it turns out mine was one of just two trios in their pilot!)
I was even more excited when the data arrived last week.
A Sneak Peek at my Results
While a more detailed analysis will be forthcoming, I’ve already had some fun diving into my exome.
Each sample arrives in an encrypted bundle, with the decryption keys stored in your 23andMe account. That bundle of data, weighing in at around 10GB, is not intended for casual use.
It contains three things:
- A PDF report, giving you a high level overview of the variants called on your exome, and a small report on the rare variants that fall within genes with known Mendelian disorders associated with them.
- A VCF file of your variants (weighing in at a measly 7MB)
- A BAM file of your aligned sequence data (roughly 10GB), generated by a HiSeq2000 and the Agilent exome capture kit.
Dr. Jung Choi recently blogged about the 23andMe variant overview PDF, so I’ll refer to his description of this report.
I’m sure many other recipients have been browsing through their report and a few may even be diving into their raw variant list.
But something I know I wanted to do for this post, something I know others receiving their exome data simply cannot, was to utilize that largest component of my exome data download: the actual sequence data in the BAM file.
Stage Right: Enter Golden Helix GenomeBrowse™
The Product Development team at Golden Helix has spent a good chunk of the last year heads down on something we think might just change the game when it comes to visualizing genomic data.
It was built from the ground up on the principles of superb user experience and on a foundation of solid engineering.
So, naturally, the first thing I did when I saw that BAM file show up in my data folder was to double-click the file and watch Golden Helix GenomeBrowse™ start rendering my exome.
If you have ever installed Google Earth, you may have experienced the addictive thrill of taking the view of the entire earth as globe and smoothly scrolling your mouse wheel while you strategically adjust the exact point in which you fly from outer space down to your own back yard.
Let me tell ya, it’s more addictive when you’re flying from a list of chromosomes, to a single arm of chromosome 10, down past the chromosome banding stains, through a gene cluster and into an exon of your own gene that has an interesting highlighted variant. And then, intuitively, you click and grab that plot to pan around.
It’s just… fun.
But just like staring at a globe and wondering where to zoom next, you quickly decide it might make sense to type in the address you want to investigate instead of panning around the whole city looking for a street name to pop up.
In my exome PDF report, there were 14 variants that were rare heterozygous nonsynonymous mutations in genes with known clinical implications.
Here is an example of the first variant (and in fact, one of the most interesting):
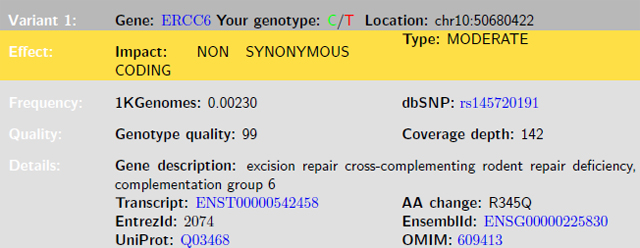
What’s immediately important about this variant is that it changes one amino acid in the transcribed protein this gene encodes for, it’s quite rare, and it occurs in a gene linked with severe genetic disorders.
Here is what it looks like when I type in chr10:50680422 into the GenomeBrowse location bar:

Reading up on ERCC6 in the OMIM database (a repository of publications related to Mendelian disease and the genes implicated), it looks like ERCC6 is an important gene involved in DNA repair and gene regulation. It has been associated with Age Related Macular Degeneration (ARMD), UV light sensitivity, and a severe rare disease causing skeletal and developmental issues called Cockayne syndrome.
By looking into the details of these published findings, it’s clear that ERCC6 is haplosufficient, as there were a number of reports of carriers of the studied mutations being unaffected.
But how likely is it that this C to T mutation actually does “knock out” or inactivate one of my two copies of ERCC6? Well, I used SVS to annotate this variant with various public databases and tools to find out.
| 1kG Overal Freq | European Freq | Asian Freq | African Freq | NHLBI Freq | NHLBI 6500 Genotype Counts |
| 0.0023 | 0.0026 | 0 | 0 | 0.001461 | TT=0/TC=19/CC=6484 |
Frequency information for chr10:50680422 C/T
| HGVS Protein | PolyPhen2 Score | PolyPhen2 Prediction | GERP++ RS | PhyloP | PhastCons |
| ERCC6 p.Arg975Gln | 0.71 | Probably-Damaging | 5.56 | 2.771 | 1.0 |
Functional Prediction and Conservation of chr10:50680422 C/T
It turns out this variant is not only rare, but present only in Europeans. Its frequency is even lower in the NHLBI Exome Sequencing Project than in the 1000 Genomes sample set. Out of the 6,503 individuals from the NHLBI Exome Sequencing Project, not a single one has this variant in both copies of ERCC6 (as a homozgyous variant) and only 19 have it in one copy of ERCC6.
Besides it just being rare, there are tools such as PolyPhen2 to measure how likely a given mutation is to damage or make inoperable the protein encoded by a gene. These tools mine databases of protein structure information and also look at how well conserved a given amino acid is across all the species that share a given gene. GERP++ Rejected Substitutions scores, PhyloP, and PhastCons are all measurements of conservation as well.
In summary, there is a good chance ERCC6 p.Arg975Gln is indeed a damaging mutation, and it’s probably a good thing my wife is not also a carrier of this variant.
Rare Variant? Rare in Which Population?
When annotating my variants in SVS, I noticed something about the 14 variants 23andMe had prioritized. Many of them, like chr10:50680422 C/T, where unique to Europeans.
In fact, if I were to use the allele frequency within just the 379 Europeans from the 1000 Genome project, half of the 14 “rare” variants have an allele frequency greater than or equal to 1%.
I once heard at a conference by a population geneticist: All variants are common in some sub-population somewhere on the globe.
So, when prioritizing the variants in my exome to investigate, I ranked them by their European allele frequencies and secondly by the annotations from SIFT, PolyPhen2, GERP++, and PhyloP. Luckily, all of these sources are aggregated in the dbNSFP 2.0 track we just added to SVS.
After ERCC6, a variant in the gene STAR came up next.

Reading through the OMIM summary, it’s clear that like ERCC6, STAR is haplosufficient such that carriers of a single mutation are asymptomatic.
Reading about which very specific organs this gene is found to be active in, and what vital hormones it’s involved in regulating, I found myself a bit unsettled in my decision to be blogging about it. Let’s just say my manhood is hostage to those 20 measly reads that show I have at least one working copy of this gene.
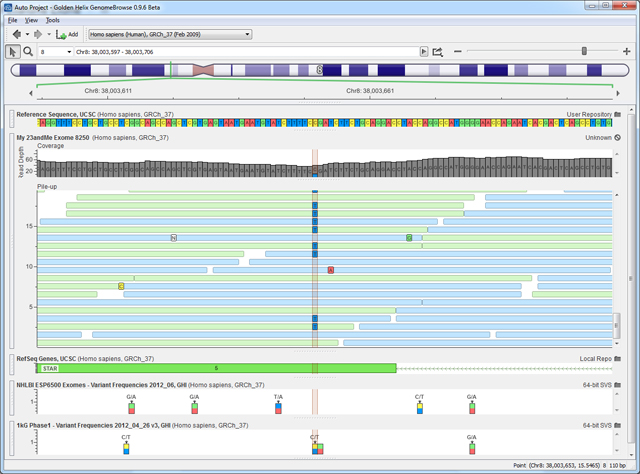
If fact, the assumption that my C>T mutations is damaging to STAR is far from certain. The PolyPhen2 prediction is that the mutation is “Benign”, yet it is at a highly conserved locus.
In a clinical diagnostic setting, this bioinformatic information would simply motivate a more definitive orthogonal test if we had worked down to a short list of putative causal variants. For example, a western blot assay could be done to validate the reduction or annihilation of the STAR protein product in the source tissue.

So much information!
While it’s fun browsing my exome, as I knew going into this, there is only so much analysis to dream up with a single healthy individual.
Over the next couple of weeks, when I have a few hours to spare, I’ll be taking a deeper dive into my full trio.
With the possibility of finding recessively inherited variants, looking for de Novo mutations, or playing with ways to interpret the genetic model of a complex autoimmune disease in the context of a single exome, I’m sure to have many interesting things to report.
Stay tuned!
P.S. If you are interested in being notified when GenomeBrowse is released, sign up on goldenhelix.com. If you would like to actively participate in the early access program, email me!

Cool. I really want to dive into genotyping my own DNA as well. And perhaps my fathers and mothers. That’ll be cool. First just the regular genotyping. But a 1000 dollar to sequence, can’t wait till that becomes really cheap!
Nice post, and it’s truly exciting you were able to get the exome trio of your family. Your GenomeBrowse looks great! I noticed that your 23andMe gene annotation incorrectly identifies the changed amino acid as R345Q for ERCC6, instead of the correct R975Q. This is a frequent issue in my gene reports from 23andMe, which has some completely inaccurate calls. It looks like your GenomeBrowse will do a much better job.
@Gabe
Thanks for posting this, we’re really excited at 23andMe to hear how people are using their data. One thing to be aware of when looking at the BAM file is that in some cases it might not match up exactly to the VCF. The BAM that you have contains all of your data, however we subsequently took the subset of this BAM that aligns on or near the target exons and reprocessed it using GATK to realign around indels and recalibrate quality scores before generating the VCF.
@Jung
The difference in the amino acid coordinates that you see is due to the presence of alternative transcription start sites for this gene. Ensembl has a nice page that shows you the amino acid coordinates for this variant in all the proteins produced by this gene: http://goo.gl/swjFY. In order to keep the VCF manageable we only report the highest impact effect for each variant (as determined by snpEff and GATK). The majority of human genes generate some form of alternative transcript (using alternative transcription start sites, alternative polyA sites or alternative splicing) so this is probably why many of the reported amino acid coordinates don’t match up.
Jung: Thanks for the comment! I sent you early access details for GenomeBrowse. Let me know if you have any issue using it for your own exome.
Eoghan is right though. On your report and mine, the amino acid change reported is not “wrong”, merely not based on the same transcript as you found using another tool.
Ensembl as a gene annotation source has the most prolific list of alternative splicing transcripts for a given gene. I annotated by variants in SVS against RefSeqGenes, as they have just the most common transcripts in their gene annotations and are often the source used when describing variants in papers. Technically you should also include the transcript ID when reporting AA mutations. Such as NM_000124 p.Arg975Gln or ENST00000355832 p.Arg975Gln.
So 23andMe is not wrong because they reported the transcript in the report that AA change is based on (to the left of the AA change). On the other hand, they are a bit wrong as it’s strange which transcript those choose to report. While I’m sure they always report the most damaging transcript interaction, there is often many transcript in which the variant is non-synonymous and yet you still need to pick one to report!
Ensembl, deep in their databases somewhere does keep track of the “cononical” transcript for a gene. In this case, it’s the one shared with RefSeqGenes and CCDS (another gene annotation database) and it’s strange that the tool 23andMe used to do variant classification didn’t pick it as well. What makes a “canonical” transcript is a debate in itself, and failing all else, Ensembl picks the one with the longest protein-coding product.
I’ve been thinking about doing a post on these confounding issues of gene and transcript usage in literature, and this is just another example of the confusion it can cause.
Really cool that you got all that information and were “able to play” around with it. It is obvious that you know what you are doing and talking about. However, in the direct to consumer model that 23andMe uses, how is the average person going to 1) understand, 2) interpret and 3) use all this information. Most of the public has a had enough time understanding extra or missing chromosomes, if they even know what they are. As far as your son, what if you found a de novo mutation that increases his risk for something later onset like Huntington, or schizophrenia? In your example of diabetes or coronary artery diseases most genetic changes are linked to a susceptibility loci and do not definitely mean you will develop the disease. While, diet and monitoring to catch something earlier may be proactive steps you can take, it is something everyone should be doing anyway. Why obsess about something that “could” happen. I “could” get hit by a bus today, does that mean I should not go outside?
Nice read! I enjoyed Jung’s blog entry on exome annotation before, but only found yours today. What is your experience with indels and the “final” release of the 23andMe pilot exome calls (something which myself and Eoghan briefly discussed on 23andMe’s boards?)
Hey Humilitan,
I didn’t participate in the 23andMe board discussion, but did email 23andMe directly right after getting the “final” release showcases the issue with bogus indel calls. I wanted to give them a chance to respond before blogging about it. Turns out it was a bug in an updated GATK version that went away with yet another GATK update. I talked to them about it some more in person at ASHG.
Given GATK is potentially the most widely used tool in the research community for this work, it definitely is shocking to hear how easy it is to run into such a serious bug.
Pingback: GATK is a Research Tool. Clinics Beware. | Our 2 SNPs…®