A common question that comes through support is if there are options in SVS for doing gender inference or checks. There is indeed functionality in SVS for this QC check! This function is under the Genotype Menu for Sample Statistics; there are a lot of great statistics available to check the quality of your data in SVS, but I’ll walk you through the Gender Inference tool.
This functionality takes your specified sex chromosome and calculates the heterozygosity rate, and based on the desired threshold will provide a call for the inferred gender per sample. This functionality is also designed to work for animals with a ZW sex determination system, such as chickens and some species of fish and reptiles. Figure 1 shows the menu options for a chicken GWAS dataset with the Z Chromosome selected for gender inference.
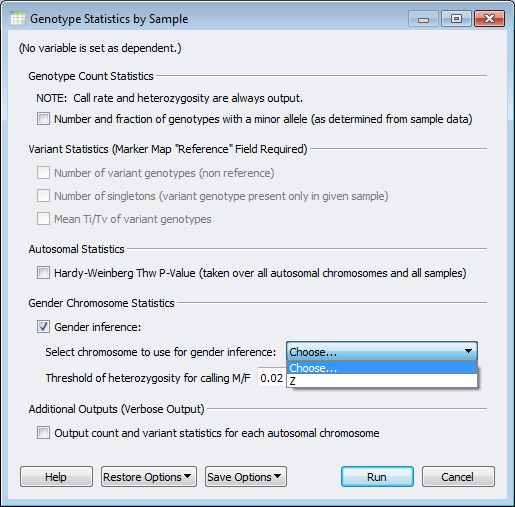
Figure 1. Genotype Statistics by Sample with Chicken Z Chromosome selected for inference
In humans, and all other mammals, females are the homogametic sex and we use the X chromosome for inference with a threshold of 0.2 being acceptable. After running this function, you can see in Figure 2 below a normal distribution for the female heterozygous rate, but the males have hardly any heterozygosity, which we would expect due to human males being hemizygous or the heterogametic sex.
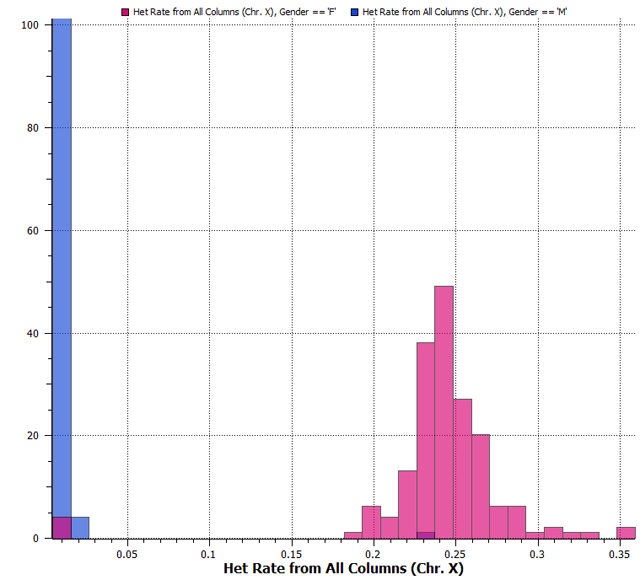 Figure 2. Heterozygosity rate histogram colored by reported gender.
Figure 2. Heterozygosity rate histogram colored by reported gender.
You can see in Figure 2, there are areas in purple which indicate samples that were reported the opposite gender than their X heterozygosity rate indicates, which could be due to a reporting error or potential sample contamination. At this point you can check the original records or decide to remove these samples before continuing your analysis.
SVS has many other options for performing quality assurance for your GWAS and DNASeq datasets; I’ve merely highlighted one option here that is frequently asked about. If you have any questions about what is possible in SVS please don’t hesitate to contact our Support Team at [email protected].