Speaking as somebody with a long history in data analysis, there are few things I find more exciting and tantalizing than new analysis methods that might apply to a problem I am trying to solve or was unable to solve in the past. Whenever I make a breakthrough in one project, I find I want to abandon the current project and go apply the new approach to an old project to see if I can make progress where there was previously a dead end. I think this is a common trait among analysts and even among scientists in general. We are naturally curious and don’t like to leave any question unanswered.
It is therefore not surprising that whenever Golden Helix introduces new features in the SNP & Variation Suite (SVS) software package we immediately get questions from researchers who want to know how the features might apply to their specific situation, even if it may seem unrelated on the surface. In recent months we have received numerous questions about how our new sequence analysis tools might apply to data from standard GWAS arrays. This is a very interesting question, and there are numerous potential applications for sequence tools in GWAS data.
Today I will demonstrate how two components of the SVS Sequence Analysis module – annotation-based filtering and variant map visualizations – can be applied to data from standard genotyping arrays.
Annotation-based filtering
In sequence analysis, annotation-based filtering is used in aspects of both data quality assurance (QA) and analysis. From a QA perspective, public annotation data can be used to identify genomic regions susceptible to poor sequence alignment you may wish to omit from the analysis. From an analysis perspective, annotation data can be used to reduce sequence data to regions of interest or to identify a subset of variants with particularly interesting characteristics. Is there any corollary for such functions in GWAS data? Yes.
One problem I have often encountered in CNV GWAS and, to a lesser degree, in SNP GWAS, is confounding as a result of segmental duplications. When reviewing the results of a CNV study, it is always a good idea to check that none of the significant results are in regions of known segmental duplications. For example, there is a small block of markers on the Affymetrix 6.0 array that are mapped to chromosome 1, but fall within a region that has high homology to a region of chromosome X. This region will invariably appear to have intensity differences that correlate with gender, which can in turn confound the results of an analysis where the phenotype is also correlated with gender. To prevent such confounding from the onset, you might wish to use filtering tools to remove markers that fall within segmental duplications from your data before starting the CNV segmentation process.
To demonstrate, I downloaded the “genomicSuperDups” table (“Duplications of >1000 Bases of Non-RepeatMasked Sequence”) from the UCSC genome browser and imported it into SVS as a spreadsheet. The table contains a listing of all genomic loci at least 1 kb in length with at least 90% similarity to at least one other locus. I proceeded to generate several annotation tracks based on that table, including tracks for all loci with >90% similarity to another region, >95% similarity, loci with >99% similarity, and any locus with at least 90% similarity to segments found on the X or Y chromosomes. I then compared these tracks to the Affymetrix 6.0 and Illumina Omni1-Quad arrays using annotation-based filtering to find out if any markers on either array might be in a questionable position.
Table: Markers from 2 common GWAS arrays located in known segmental duplications, based on UCSC annotations.
| Affymetrix 6.0 (na30 map) | Illumina Omni1-Quad | |
| Total Autosome SNP/CN Markers | 1,778,648 | 1,109,421 |
| Autosome markers in segment dups of at least 90% similarity to regions on any chromosome | 41,702 (2.3%) | 52,471 (4.7%) |
| Autosome markers in segment dups of at least 95% similarity to regions on any chromosome | 23,873 (1.3%) | 33,787 (3.0%) |
| Autosome markers in segment dups of at least 99% similarity to regions on any chromosome | 4,024 (0.2%) | 12,847 (1.2%) |
| Autosome markers in segment dups of at least 90% similarity to regions of X or Y chromosome | 1989 (0.1%) | 3330 (0.3%) |
We can clearly see from the table that both arrays include substantial content from within segmental duplications. Of course, the human genome is known to be highly repetitive, and it is difficult to avoid such regions in designing a true genome-wide assay. The array manufacturers consider sequence identity in the probe design process, and we can assume most of the markers in these regions are based on unique sequences. However, there remains a risk of non-specific binding for some probes, which at worst might lead to false-positive CNV identification and at best might result in a bit of data noise. If you are concerned about this possibility, or believe it may have already affected your results, annotation-based filtering provides an easy solution to remove such regions in advance of analysis, or even to inactivate them in a p-value spreadsheet after analysis.
The concept of filtering segmental duplication regions out of CNV data is just one of many possible applications of annotation-based filtering with GWAS data. In SVS, most of the filtering tools can be used on any marker-mapped spreadsheet, whether the map is applied to rows or columns. So if you want to filter a P-value spreadsheet to show only results from exonic SNPs, or to hide results within 50 bp of an STR region, you can do so. In a closely-related item for SVS users, the latest update (v7.5.4) includes a new feature called “Set Genotypes to Missing Based on Second Spreadsheet.” This tool was designed to filter sequence variant calls based on a read depth spreadsheet, but it could also be used in GWAS for tasks like filtering genotypes based on CNV data from a spreadsheet of Segmentation Covariates or Log Ratios.
The possibilities of filtering are limited only by your imagination.
Variant Map Visualization
Sequence variant map visualization in SVS is a powerful way to interactively explore sequence variants. It has the capability of visualizing SNVs, substitutions, and indels for any number of subjects simultaneously. The content of most genotyping arrays is limited to SNPs, but there is no reason why you can’t still use the variant map function to visualize the SNP data if you so desire. This type of visualization can be much more informative than reading a spreadsheet when trying to assess broad patterns of variation. The one requirement for drawing a variant map is that the marker map associated with the genotype spreadsheet contains a field for the reference allele. The allele coding for the SNPs doesn’t even need to come from the same strand as the reference allele for the visualization to work, although you will find the visualization is more useful when all data is coming from the same strand.
Let’s consider an example using Affymetrix 500k data for HapMap subjects. For this example, I started from a spreadsheet containing genotypes coded according to the Affymetrix “Top Alleles“ definition (which happens to be a reasonable match for the human reference genome). The associated marker map does not contain a reference allele field, but I can add one using either of two workflows in SVS. First, to simply define the reference as the major allele, I could run “Genotype Statistics by Marker” and then add the major allele column to the marker map. Using the major allele as a pseudo-reference works nicely for visualization, but any comparisons to annotation tracks may be difficult to contextualize. I chose to use a more advanced workflow, and defined the reference allele based on the actual NCBI36 reference sequence. (For step-by-step instructions, feel free to contact support or check out Autumn’s recent blog post about adding data to a marker map.)
Once the reference allele field has been added to the marker map, I am ready to make a variant map. The figure below shows the final result in the SVS genome browser, zoomed to the area around the gene MAP3K5.
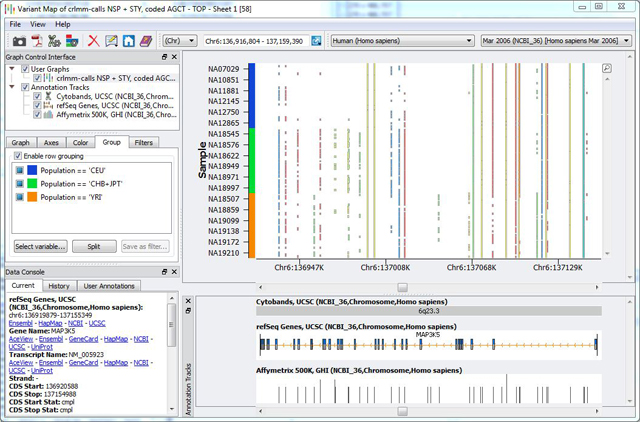
Figure: The SVS Genome Browser showing a variant map for 270 HapMap subjects. The three population groups are identified by the colors of the Y axis. This visualization is typically used for sequence variants identified with NGS technologies, but is used here to show genotypes from the Affymetrix 500k array.
From this view, you can immediately identify patterns of variation unique to each of the three population groups. Remember that the allele coloring only requires that one allele be different from the specified reference, so it is not obvious which points are heterozygous and homozygous, but this visualization is still useful for a high-level exploration of variation patterns.
Other Tools in the SVS Sequence Analysis Module
On the surface, it might seem like all of the SVS Sequence Analysis module tools can be used for analyzing array data, and indeed, there is nothing in the software to prevent you from doing so. But there are a few caveats to be aware of. These issues pertain to almost all sequence analysis tools – not just SVS.
Among the most important issues is the question of genotype strand, which was mentioned in the variant map discussion above. Some of the most powerful tools in the Sequence Analysis module (including the CMC test, Variant Annotations, SIFT filtering, etc.) work on the assumption that all variants have been called based on the published sequence of one of the standard reference genomes. Unfortunately, the default genotype calling algorithms for Illumina and Affymetrix arrays don’t necessarily adhere to that standard. For most arrays, the data is available to help you translate your data to the right strand, but you can’t assume it is going to match up correctly in advance, particularly with older arrays.
Another big issue relates to data quality for rare variants. The Sequence Analysis module is developed with a primary purpose of enabling rare variant analysis using tools like the KBAC and CMC collapsing tests. The new generation of genotyping arrays, including the Illumina Omni5 and the forthcoming exome chip, have extensive rare content. I expect the rare variant tests will be very useful for the data from these platforms, just so long as the genotype calls are accurate. For most of the GWAS era, researchers have habitually filtered low-frequency SNPs from their data, usually with a threshold between 1% and 5%. This practice is due partially to the low statistical power associated with rare alleles, but also due to unreliable genotype calling. I have heard repeatedly the technology of the new arrays has improved, and the rare allele calls can be trusted with higher certainty. I hope this is true, and I look forward to the opportunity to work with data from these new platforms.
That’s It!
So there you have it. SVS is designed to be very flexible, and you should never feel constrained by the workflows presented in the tutorials. We are often surprised to learn about the ways in which our customers use the software. We would love to hear your feedback about the unique applications you have found for SVS. I hope these two examples of non-standard applications will help you to see some of the possibilities for applying new analysis tools to your data.
…And that’s my 2 SNPs.