A new VSPipeline command, set_data_folder_path, designed to bolster consistent input usage. By introducing this innovative command, we aim to empower users with improved data organization, flexibility, and standardization for their clinical cases and analyses. Embracing this command will not only support reproducibility but also ensure accountability, ultimately paving the way for better-informed patient care decisions.
Managing Annotations and References in VSPipeline
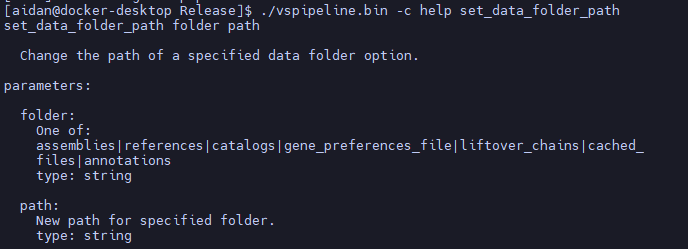
Reproducibility is a cornerstone of robust clinical genomics and a key principle of Golden Helix Software. The project templates used by VarSeq and VSPipeline allow saving a set of parameterized algorithms and should be your first stop when setting up a reproducible pipeline. Besides using templates for the algorithm settings, reproducibility requires ensuring consistent inputs. This article discusses the new VSPipeline command: set_data_folder_path, which allows users to specify the folder of reference samples and other supporting inputs to streamline reproducibility in your clinical genomics pipeline.
Overview of the ‘set_data_folder_path’ Command Line Argument
There are several different paths that can be set with this command:
- assemblies – The folder where the genome assemblies are located.
- references – The folder where the CNV references are located.
- catalogs – The folder where the assessment catalogs are located.
- gene_preference_file – The path to the user gene preferences file, used by vsclinical and the annotate transcripts algorithm
- liftover_chains – The folder with the chain files to use if you have liftover as part of the import pipeline.
- cached_files – The folder for cached files when reading network files
- annotations – The folder to look in for annotations and other supporting inputs for the algorithms in the VSProject template.
Managing Gene Preferences
The set_data_folder_path gene_preference_file <file> allows users to set the path to a file containing gene-specific settings. The gene_preferences file includes information such as the default transcript, disorder, and the disorder inheritance model for specific genes; more information about this file and how it is used can be found in the manual.
By incorporating the ‘gene_preferences’ flag in the analysis, clinical genomics professionals can ensure that the same gene-specific settings are applied consistently across different cases and analyses. Additionally, it allows for different preferences to be used for different pipelines; for example, you may want to use different transcripts depending on the tissue type of the sample or the disorder that you are screening for.
Managing CNVs Reference Files
The set_data_folder_path references <path> command enables defining the folder of reference samples used for CNV detection. Using this command in your vs-batch files has several advantages:
- Organization: By allowing users to set a specific folder for reference samples, ‘set_data_folder_path’ promotes better organization of data, making it easier to manage and maintain consistency in clinical genomics workflows.
- Flexibility: Clinical genomics professionals can quickly switch between different sets of reference samples by changing the folder path, enabling them to work with various datasets and compare results across different reference sample sets.
- Accountability: The ‘set_data_folder_path’ argument helps ensure that all vspipeline runs use the same reference samples, contributing to reproducibility and consistency in a clinical setting, and specifying it in the vs-pipeline batch script makes it explicit which reference folder was used.
As the quality of the reference samples is an important factor in the final results produced by the CNV caller. This new option offers better control to track and organize your reference samples and an easy way to switch between reference sets if your sample prep changes.
Conclusion
The set_data_folder_path command line argument is a valuable tool for enhancing reproducibility in clinical genomics by streamlining the use of annotations and reference samples. By allowing users to define folders with the inputs to use for each run, this feature promotes better organization, flexibility, and standardization across clinical cases and analyses. This, in turn, will promote accountability, support reproducibility in the field, and ultimately lead to better-informed clinical decisions and patient care. For any questions about this new command, please contact [email protected]