In the 1990s the genetic industry voiced a request for a variant catalog that incorporates associated variant information such as phenotypic and metabolic pathways. The call was answered by NCBI, which created dbSNP; dbSNP became publicly available in 1998 and around 1.5 million variants. Fast forward to the present and dbSNP now contains over 2 billion SNPs spanning human, rat, mouse, chimpanzee, and the malarial parasite. As you may have guessed, there is an updated version of dbSNP (dbSNP 154 v2), which is now available in VarSeq.
To provide a brief overview of dbSNP, this catalog contains records for single nucleotide variants, short insertions and deletions, short tandem repeats, and microsatellites for both common and rare variants. This database also integrates resources that allow users to understand disease associations, genotypic information, and allele origin to obtain a better context of the variant. More importantly, dbSNP 154 v2 incorporates new features, such as supporting HGVS and protein variant search, RefSNP page linked to LitVar information, and incorporates the ALFA frequency for all SNPs.
dbSNP Reference SNP (RefSNP) is a locus accession for a human variant type assigned by dbSNP, commonly known as an rsID. This RefSNP catalog is a non-redundant collection of submitted variants that provides a stable notation for mutation and polymorphism analysis, annotation, reporting, data mining, and integration. This feature is now linked to LitVar.
LitVar allows the search and retrieval of relevant variant information from biomedical literature and shows key biological relations between a variant and its close related entities (e.g. genes, diseases, and drugs). The LitVar results are also automatically extracted from over 30 million PubMed articles as well as applicable full-text articles in PubMed Central. As you can see in Figure 1, by clicking on the LitVar button, you have access to all relevant literature for that particular variant.
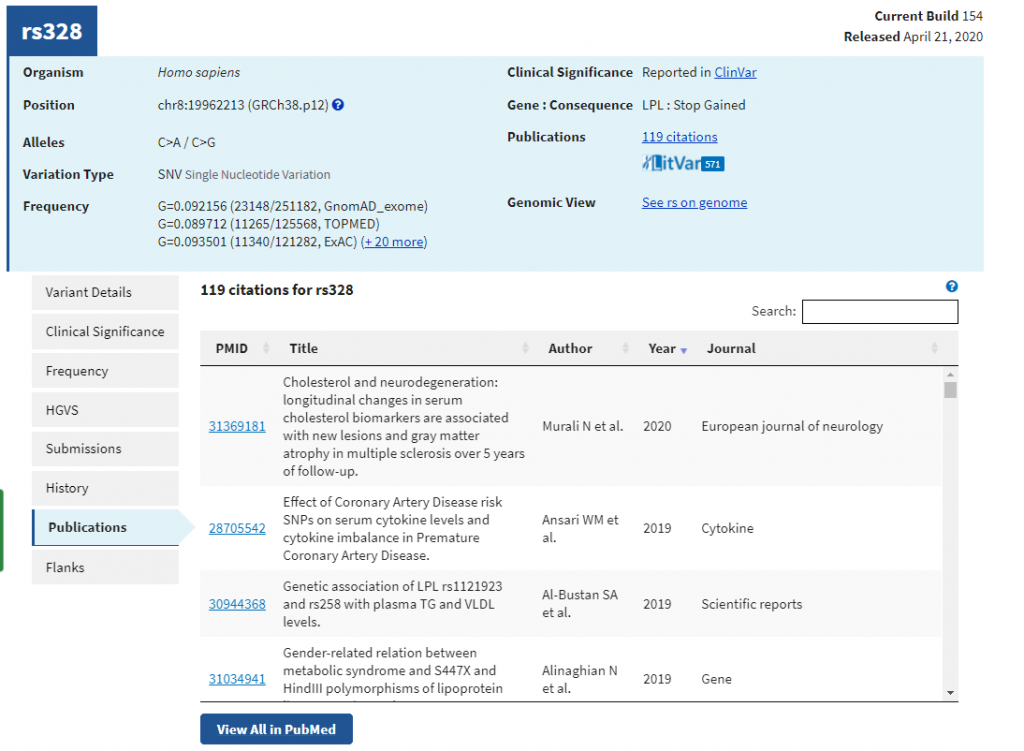
Figure 1: DbSNP 154 v2 now supports HGVS and protein variant search, RefSNP page linked to LitVar information, and incorporates the ALFA frequency for all SNPs.
DbSNP 154 v2 also incorporates the Allele Frequency Aggregator (ALFA), which was discussed in a previous blog post. In summary, this database has over 2 million subjects and hundreds of millions of variants along with thousands of phenotypes and molecular assay data. The majority of these variants were restricted to a permission basis but recently made public to promote research toward identifying variants that contribute to health and disease. As you can see in Figure 2, the ALFA frequency is one of the top frequency databases listed in dbSNP and this annotation can be integrated into your analysis as discussed in the blog.
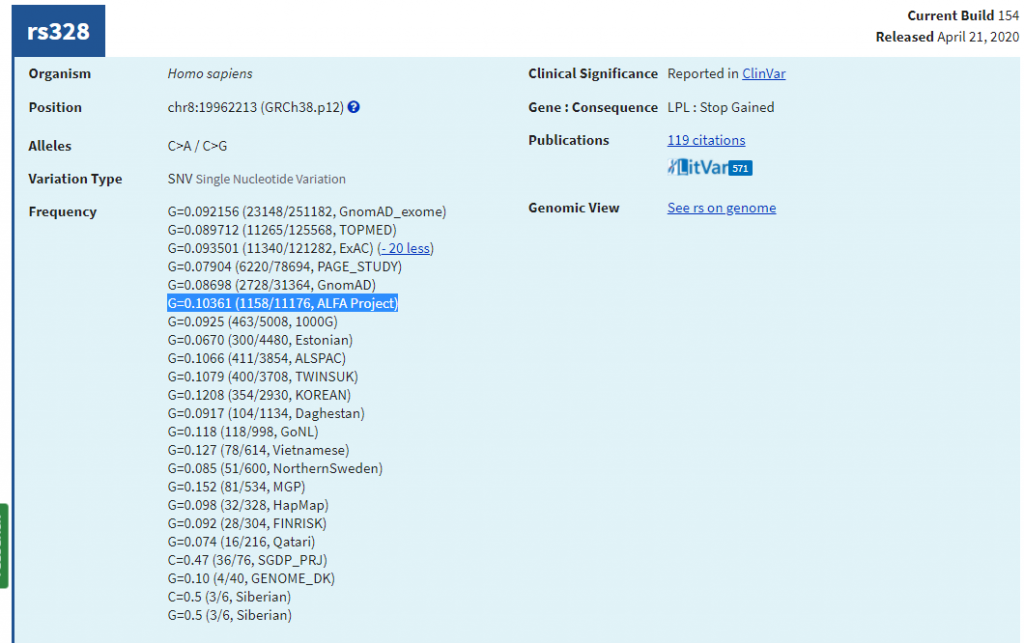
Figure 2: Allele Frequency Aggregator is available in dnSNP.
Together, dbSNP 154 v2 incorporates new features such as HGVS variant search, LitVar access and ALFA frequencies, and most importantly, this annotation is available in VarSeq for builds GRCh37 (hg19) and GRCh38 (hg38). This annotation can be added to your project by going to Add>Variant Annotation>Public Annotations, as shown in Figure 3. However, since this database contains over 2 billion genotypes, it is a relatively large annotation sitting at 12Gb. With that said, make sure you have adequate storage space if downloading locally.
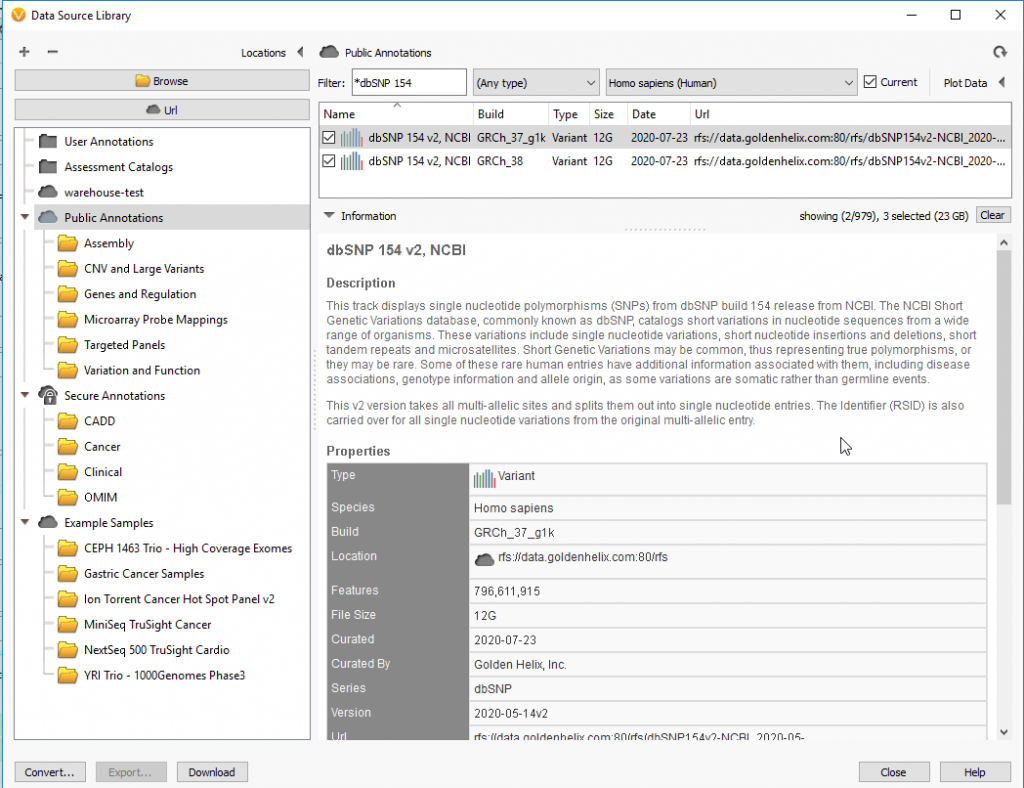
Figure 3: DbSNP 154 v2 is available in VarSeq for GRCh37 (hg19) and GRCh38 (hg38).
We hope you enjoyed reading this blog post about the new features integrated into dbSNP 154 v2 and we hope you stay tuned for more upcoming blogs and webinars. As always, thank you for reading our content and if you have any questions feel free to contact [email protected] or, if you would like to request a free trial of our software you can contact [email protected].