There are many reasons a user may wish to focus in on specific variants as part of variant annotation and filtration workflow. You may be looking for the occurrence of specific SNPs in a cohort or perhaps looking for variants known to be associated with specific forms of cancer. For both of these use cases, VarSeq provides a Match String List algorithm that is essentially a rsID lookup for use as a part of your filter chain. For those who are new to the world of NGS, a Reference SNP cluster ID (rsID) can be thought of as a barcode used to designate a particular SNP. As of VarSeq version 2.2.5, you no longer need to convert the identifier field into a string to use our rsID lookup algorithm, making this process more user-friendly. We will cover the steps for using our rsID lookup algorithm below.
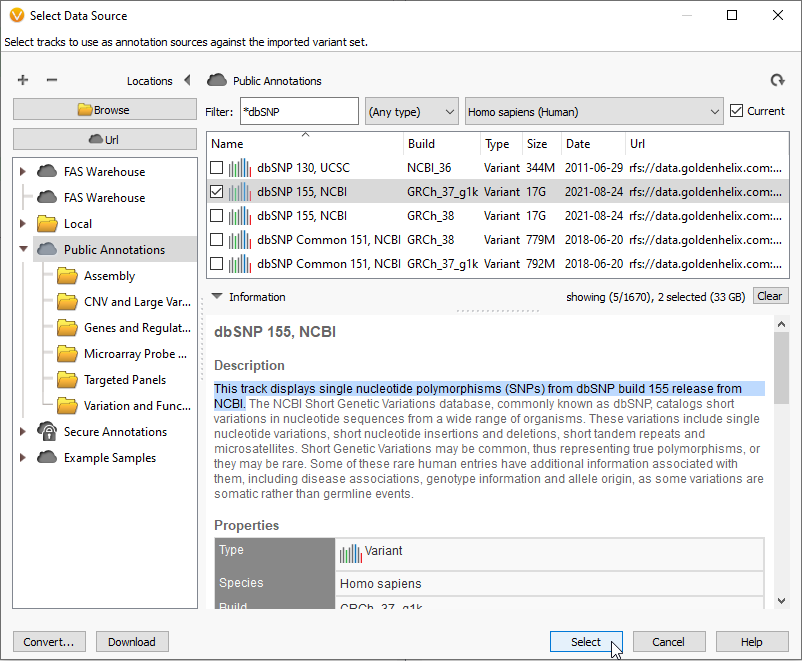
The identifier designations, or rsIDs, can be found by applying the dbSNP database as an annotation source for your project (Figure 1). dbSNP 155, for example, can be located under Public Annotations for both builds GRCh37 and GRCh38. These are large databases, over 17GB each, so check your available memory before downloading these databases for the first time.
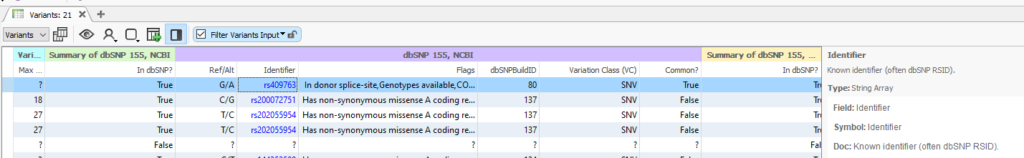
Once dbSNP has finished loading into your project, you will be able to use the rsID Identifier field with the Match String List algorithm (Figure 2). Every recorded SNP will show its own rsID, along with any Flags or notes to take into consideration. For example, this rs409763 is known to exist in a donor splice site.
To access the matching algorithms to make the best use of the rsIDs, go to Add > Computed Data (Figure 3).
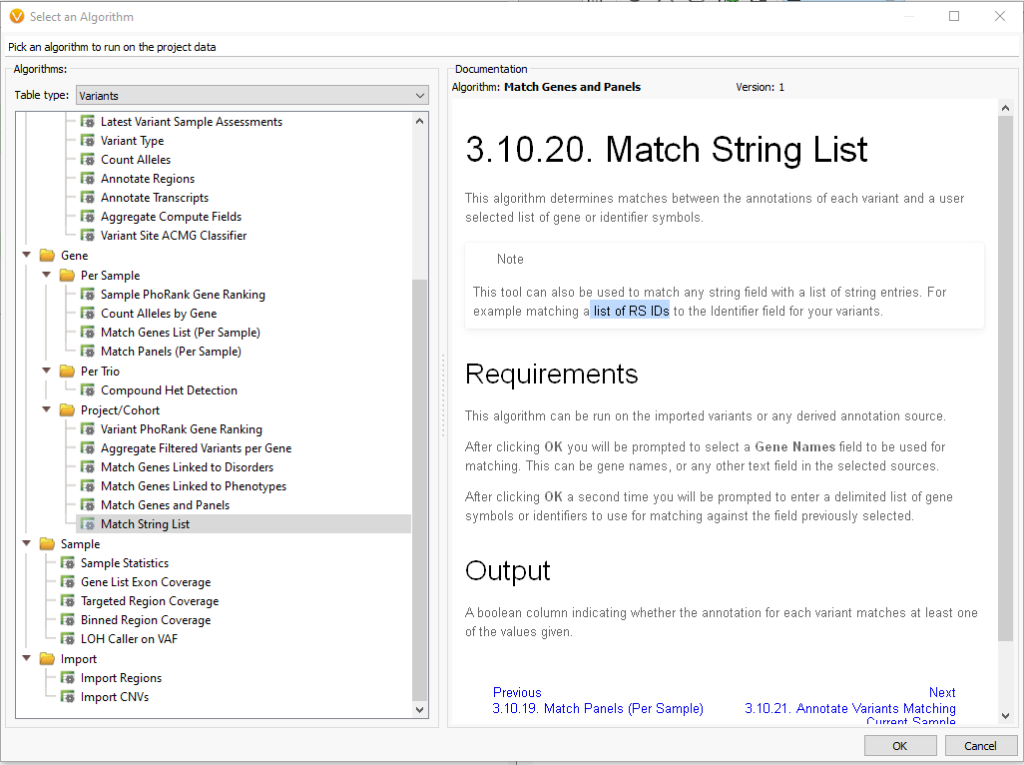
Under the Table type of Variants, there is the option to go under Gene > Project/Cohort and then access the Match String List (Figure 4). As of VarSeq 2.2.5, the Match String List algorithm accepts both Genes Lists and rsIDs.
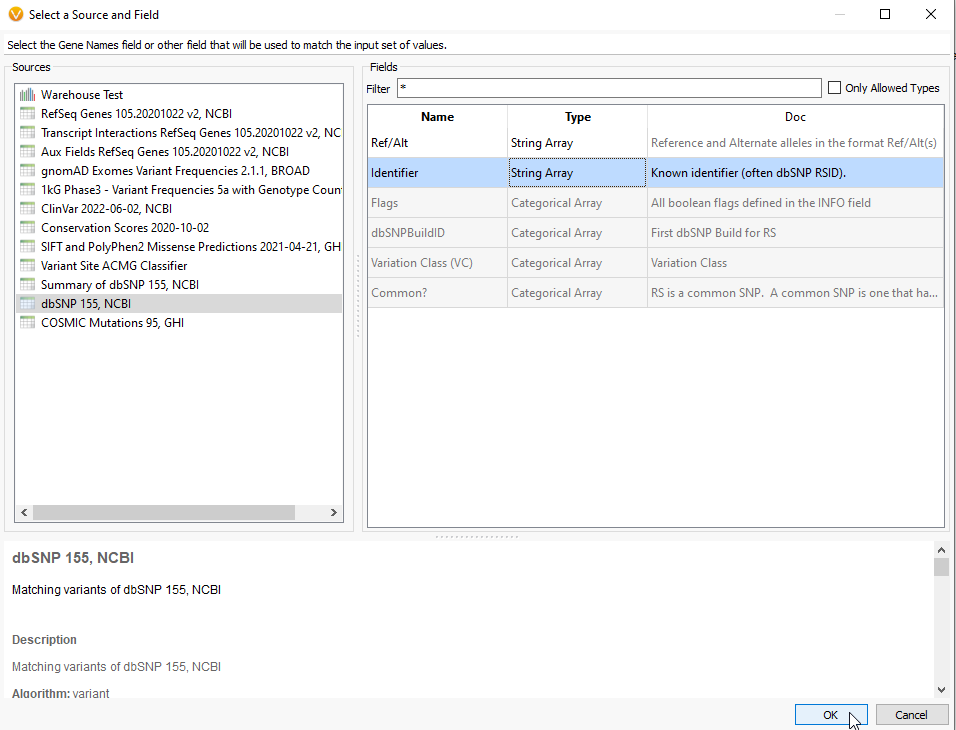
For the next panel, VarSeq will ask if you want to use Gene Names for your search. This would be appropriate for matching against a genes list, similar to running a virtual panel. For using the rsID lookup, change the source to dbSNP 155, NCBI, and select the Identifier Field (Figure 5). As you can see, a number of fields are grey, showing that they are not valid inputs for this particular algorithm. Once you have Identifier selected, hit OK.
Trial the Match String List Algorithm in VarSeq today
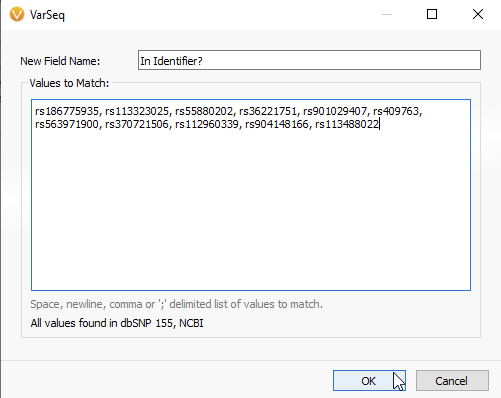
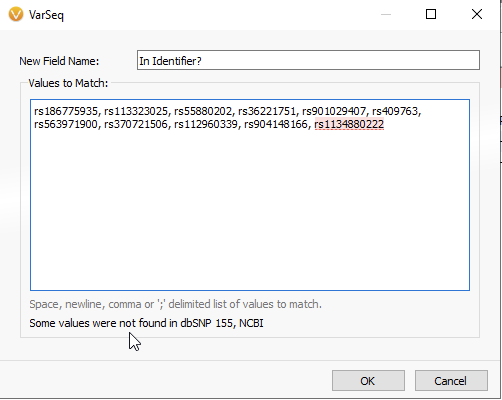
In the next panel, you will be able to copy and paste your list of rsIDs (Figure 6A). VarSeq informs you that all values have been found in the dbSNP database, while any non-matching rsIDs will be flagged red (Figure 6B). Once the list of rsIDs has been validated, you can hit OK.
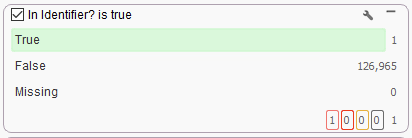
The Match String List is added as a column to the variants table, where the Match Column can be added to the filter chain for a True/False search of the available variants (Figure 7A). In this example, I have one variant with a matching rsID that makes it through my filter chain (Figure 7B).
In this blog, we covered using the improved VarSeq 2.2.5 Match String List Algorithm for bringing a rsID lookup into your workflow.
Golden Helix software provides simple, fast and repeatable variant analysis software for gene panels, exomes, and whole genomes. Our intuitive software is used for breakthroughs in cancer diagnostics, hereditary testing and diagnosis, trio analysis, and more. With integrated data visualization and unlimited support, Golden Helix’s VarSeq provides unparalleled genomic data analysis.