With the increasing knowledge of mutations involved in cancer, it is imperative to have a tertiary analysis pipeline that provides users with the most up to date information on somatic mutations. VSClinical’s Cancer Add-On does just that and more; with this feature, users can investigate and report on SNVs, indels, CNVs, gene fusions, and considerations for wild type genes in accordance with the AMP guidelines. Users can also obtain final classifications and interpretations that include drug sensitivity and resistance, as well as prognostic and diagnostic information. Also integrated into the Cancer Add-On is our Cancer Knowledge Base (CancerKB), which provides comprehensive interpretations for many common cancer genes and biomarkers for specific tumor types.
This blog will show how VSClinical’s Cancer Add-on can be used to report on a copy number amplification associated with non-small cell lung cancer (NSCLC) using the CancerKB catalog and other available resources within the platform.
Adenocarcinoma Background
Adenocarcinoma is a type of non-small cell lung cancer (NSCLC) in which the alveoli involved in the gas exchange airways of the respiratory tract become abnormal, leading to a persistent cough, shortness of breath, and potentially death. It is estimated that 1 in 15 individuals in the U.S will be diagnosed with lung cancer in their lifetime, which is concerning as lung cancer has the lowest 5-year survival rate (18%) relative to other most common cancers (1,2). That said, if lung cancer is diagnosed and treated at an early stage, the 5-year survival rate improves to 55% (1). Thus, it is important to diagnose and treat lung cancer early before it metastasizes into other areas of the body.
Lung adenocarcinoma is the result of somatic mutations primarily impacting receptor tyrosine kinase pathways. These pathways are important as they are involved in cell proliferation and survival through binding growth factors, cytokines, and hormones. Genes that are commonly mutated in lung adenocarcinoma include TP53, EGFR, KRAS and NF1(3). Additionally, copy number amplifications in oncogenes such as TERT and MET have been indicated in causing lung adenocarcinoma (3). Furthermore, diagnosis of lung adenocarcinoma is possible with genetic testing, which can also yield information for selecting a targeted therapy and potentially evaluate the suitability of a patient to be part of a clinical trial.
Case Scenario
A 50-year old female, former smoker, with cough and shortness of breath accompanied by an unintended 10-lb weight loss over a 3-month period underwent computed tomography (CT) revealing localized adenocarcinoma, indicating a type of non-small cell lung cancer, from which a biopsy was sent for genetic testing. Following genetic testing, the VCF file and BAM file were produced and imported into VarSeq.
Implementing VS-CNV & the Cancer Add-On
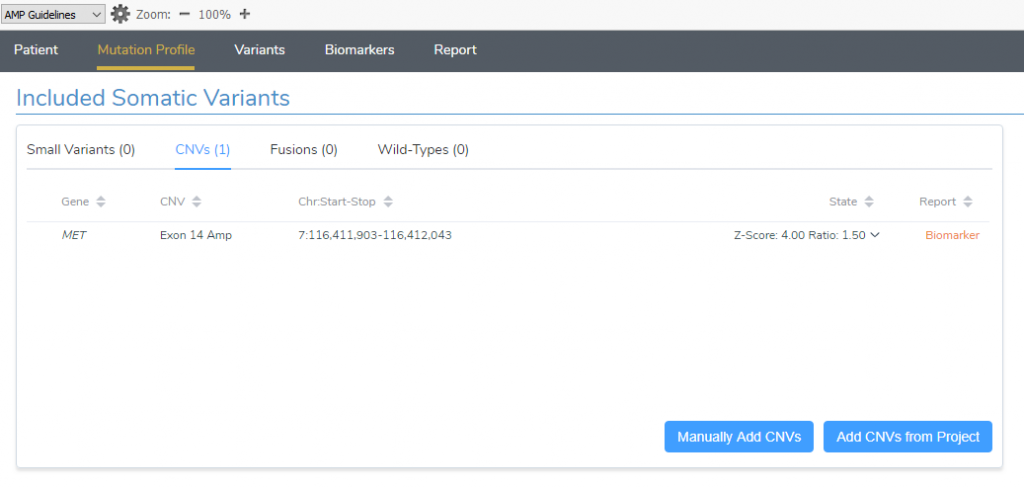
By comparing the coverage profile stored in the patients BAM file to a set of references, the VS-CNV caller identified an exon 14 amplification in the MET gene that was without sample and event quality flags. This event was added from the project into VSClinical’s Cancer Add-On and was associated with the tumor type of non-small cell lung cancer (Figure 1).
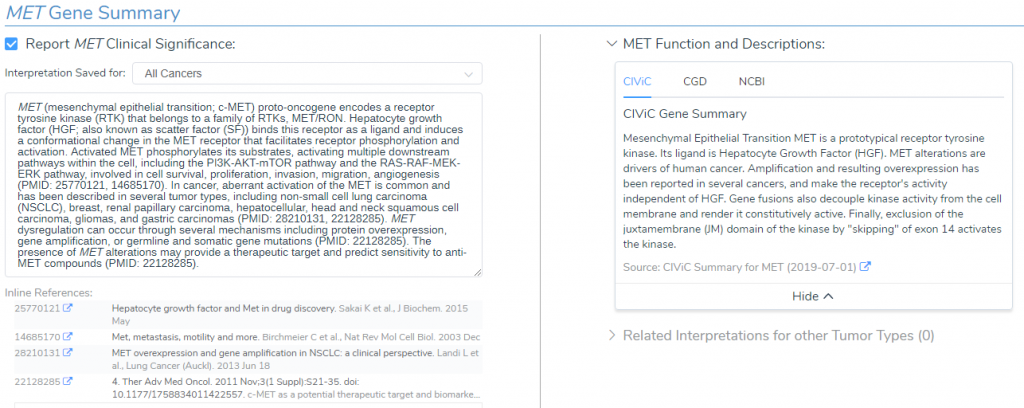
MET Gene Summary
Since the MET amplification is a common somatic mutation, the Golden Helix CancerKB catalog provided a premade interpretation for the MET gene summary (Figure 2). The summary states that the MET gene encodes a receptor tyrosine kinase that binds hepatocyte growth factor (HGF), which in turn activates downstream pathways involved in cell survival, proliferation, invasion, migration, and angiogenesis (4). Additionally, CancerKB highlights that abnormal activation of MET, via gene amplification, is common in non-small cell lung cancer and that there are potential therapeutic targets (5).
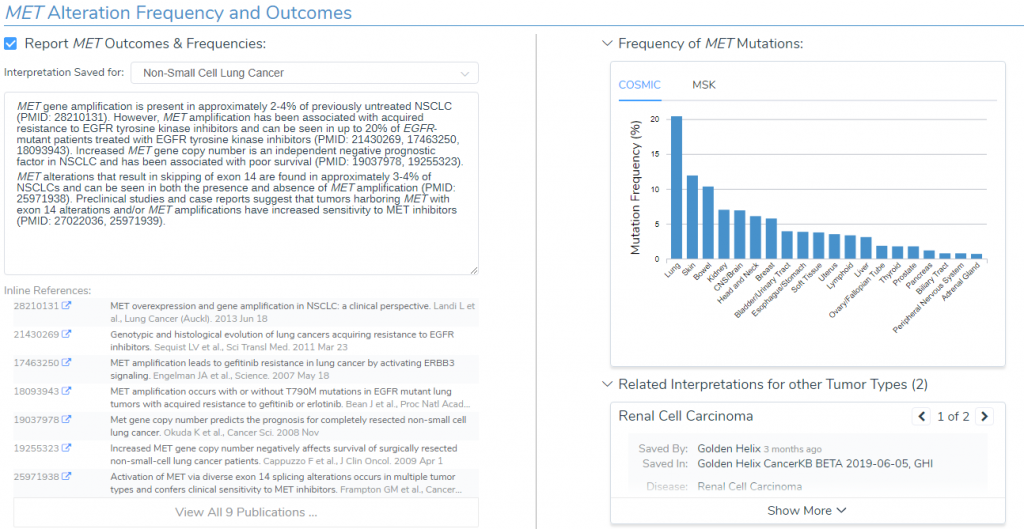
Alteration Frequency and Outcomes
CancerKB also provided detailed information regarding the frequency of somatic mutations in MET among different tissue types as well as observed outcomes (Figure 3). In summary, the lung is the primary tissue observed for somatic mutations in MET (21.58%) and the specific MET gene amplification is seen in 2-4% of previously untreated NSCLC (6). Additionally, there is resistance to EGFR tyrosine kinase inhibitors, but studies and case reports suggest that tumors harboring MET amplifications have increased sensitivity to MET inhibitors (7). Together, this indicates that there are potential therapies for the MET amplification associated with NSCLC and that EGFR tyrosine inhibitors should not be used.
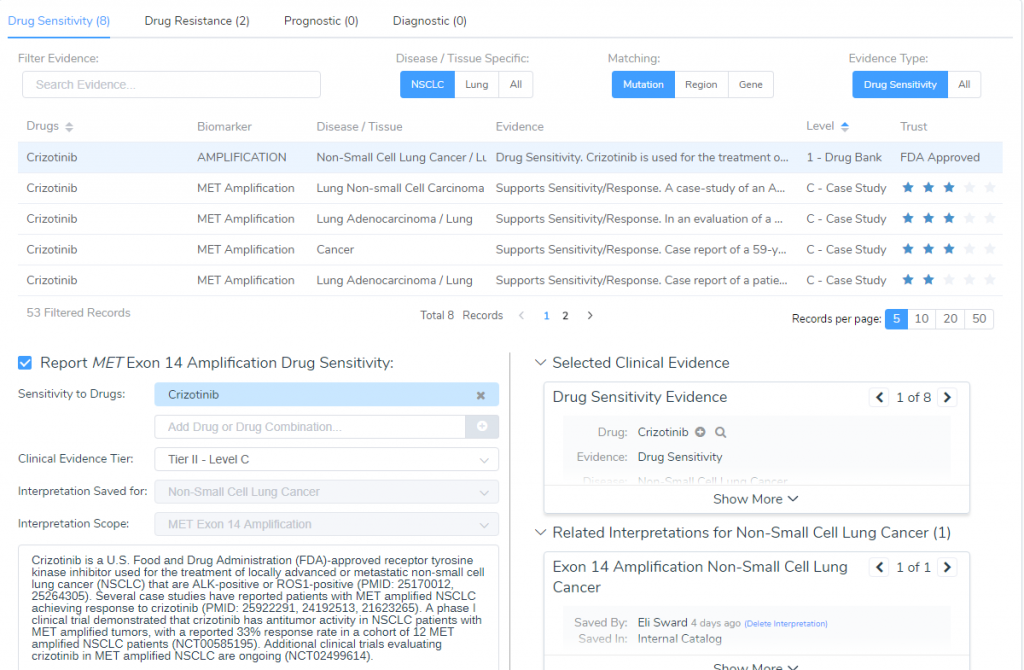
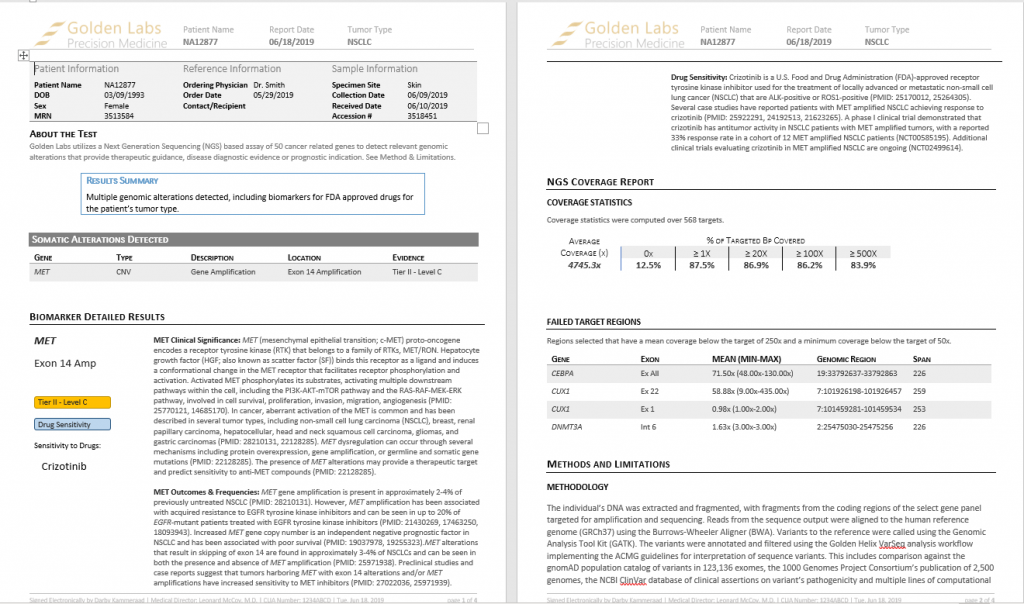
Clinical Evidence
The last step in this evaluation is determining the clinical evidence for this amplification for which Crizotinib was recommended (Figure 4). Crizotinib is an FDA-approved receptor tyrosine kinase inhibitor used for the treatment of locally advanced or metastatic NSCLC that is ALK or ROS1-positive (8). Several case studies have reported patients with MET amplified NSCLC achieving response to Crizotinib (9) and a phase I clinical trial demonstrated Crizotinib to have antitumor activity in NSCLC patients with MET amplified tumors, with a reported 33% response rate (10). Taken together, since Crizontinib is an FDA approved drug but still being tested in clinical trials, the final AMP classification for this somatic mutation is Tier-II – Level C.
Reporting
Since the MET exon 14 amplification biomarker has been interpreted, the full content of the analysis can be reviewed in the reporting section and rendered into an AMP-based clinical report (Figure 5). This report is easy to modify as it is a template based on Microsoft word, which allows you to orient the template to fit your labs specific branding needs. Most importantly, the reports can include interpretations for all variants and genes, both somatic and germline, that were added to the VSClinical interface. Lastly, all interpretations are stored in your assessment catalog, which can be utilized automatically filling in the previous findings for when you encounter the biomarker again.
Conclusion
Through VarSeq, we were able to identify an amplification in the MET gene using the coverage profile stored in the BAM file and by using VSClinical’s Cancer Add-On, we created a reportable biomarker using GoldenHelix CancerKB and identified an FDA approved drug for the cancer. Ultimately, VSClinical’s Cancer Add-On leverages a surplus of curated annotation sources and functional prediction algorithms to provide you with an extremely detailed understanding of somatic mutations. This informative yet intuitive feature, in turn, allows users to detect and report on somatic mutations and identify potential therapeutic options in the cancer workspace.

References
- https://seer.cancer.gov/statfacts/html//common.html
- http://seer.cancer.gov/statfacts/html/lungb.html
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014 Jul 31;511(7511): 543-550.
- Sakai K et al. Hepatocyte growth factor and Met in drug discovery. Journal of Biochem, 2015 May
- Sierra JR et al. c-MET as a potential therapeutic target and biomarker in cancer.Advanced Medical Oncology. PMID 22128285. 2011 Nov
- Landi Minuti et al. MET overexpression and gene amplification in NSCLC: a clinical perspective. Lung Cancer. 2013 Jun 18; 4:15-25.
- Paul K Pail, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping.Cancer Discovery. 2015 Aug; 5(8): 842-849.
- Kazandijian et al. FDA approval summary: crizotinib for treatment of metastatic non-small cell lung cancer with anaplastic lymphoma kinase rearrangements. Oncologist 2104 Oct; 19(10):5-11
- Le X, et al. Detection of Crizotinib-Sensitive Lung Adenocarcinomas with MET, ALK, and ROS1 Genomic Alterations via Comprehensive Genomic Profiling. Clinical Lung Cancer. 2015 Sep; 16(5): 105-109.
- https://clinicaltrials.gov/ct2/show/NCT00585195
Check out some of the other blogs in this series:
- Variant Interpretation with VSClinical: Clinical Example for Congenital Indifference to Pain
- Variant Interpretation with VSClinical: Evaluation of an X-linked recessive mutation
- Variant Interpretation with VSClinical: Evaluation of Hypertrophic Cardiomyopathy
- Variant Interpretation with VSClinical: Congenital Myasthenic Syndromes (CMS)
- Variant Interpretation with VSClinical: Huntington’s Disease (HD)