Variant interpretation is a critical aspect of any clinical NGS workflow. VarSeq assessment catalogs are a tool used to save variants and associated variant information for easy tracking and retrieval of completed variant interpretations. As variant interpretations stack up and classifications are saved, storing in assessment catalogs makes it easy to automatically fill in a previous interpretation if the variant were encountered again in another sample, saving the user time and effort. This blog is a simple introduction to the nature and usage of assessment catalogs in VarSeq.
VarSeq assessment catalogs save certain key pieces of information for users which vary based on the organizational schema used:
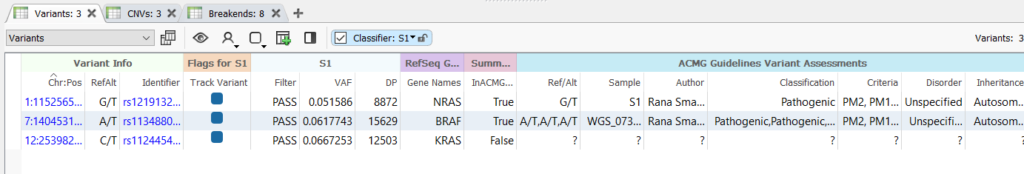
- Variant information – This includes critical fields such as the Ref/Alt and chromosome position but can also include fields like the identifier.
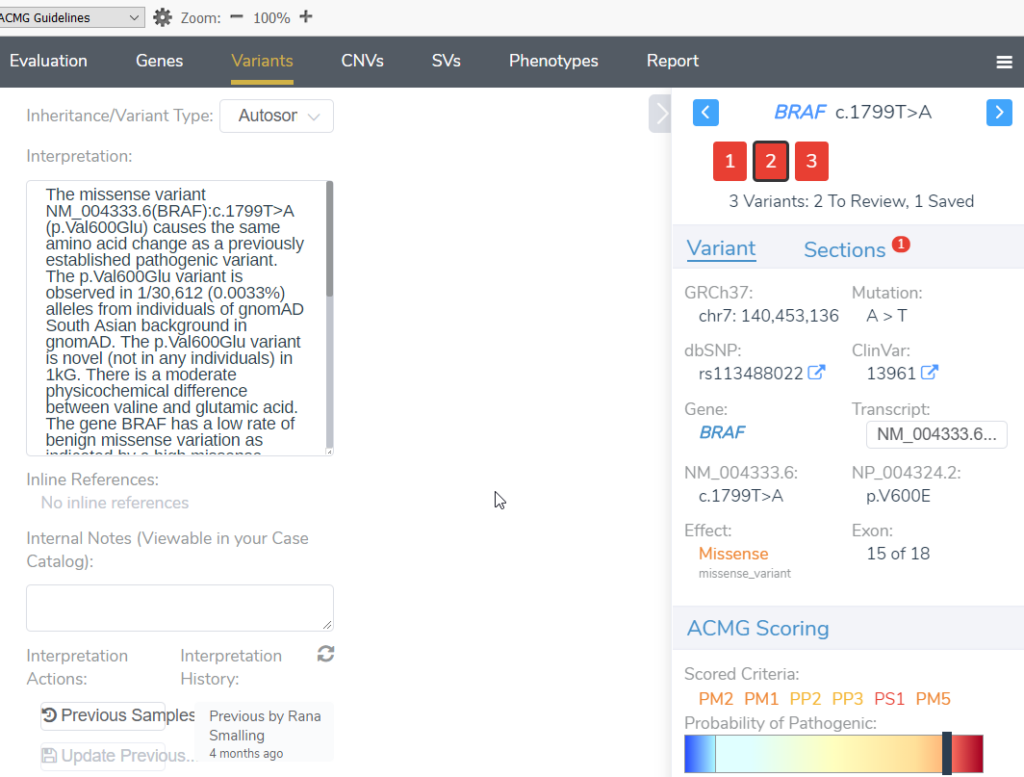
- Variant Interpretation – This is a summary of the evidence supporting a variant classification.
- Author of the interpretation – The individual user who created and saved the interpretation, and this is critical information if assessment catalogs are being shared among users on a team.
- Date of the assessment – When the assessment was created or last edited.
- Additional fields – These include various different fields based on the schema used for the catalog, for example, the disorder and notes.
How to Create an Assessment Catalog
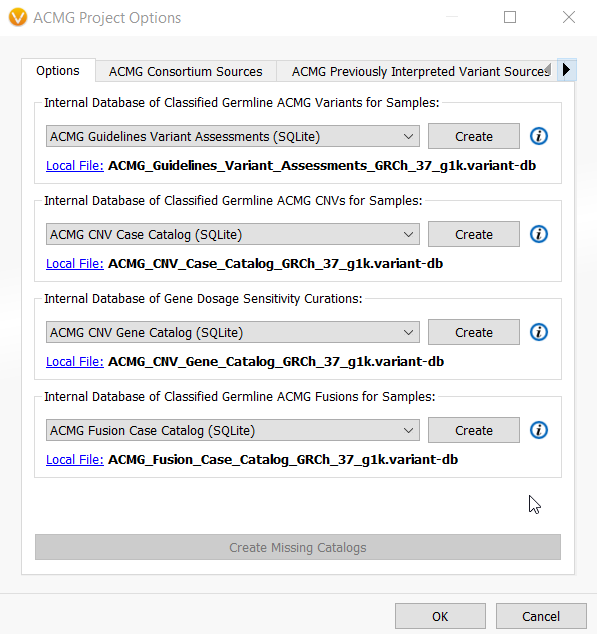
The great news is that VSClinical users will have several default assessment catalogs with well-thought-out schema created for them once they open up VSClinical and hit Create Missing Catalogs. When using the ACMG guidelines, the user will be prompted to create these default assessment catalogs for germline small variants and structural variants.
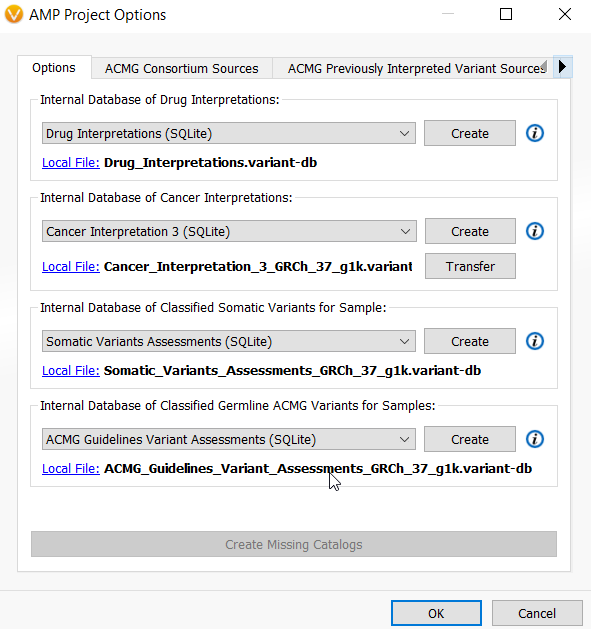
The default catalogs for the VSClinical AMP guidelines are slightly different and include a Drug Interpretations catalog for storing information on cancer drugs and a Cancer Interpretation catalog for drug sensitivity, resistance, diagnostic, prognostic, and biomarker interpretations. However, the somatic variant assessments catalogs store the usual variant interpretations, and the same ACMG Guidelines Variant Assessments catalog is reused for somatic variant analysis when storing of suspected germline variants.
Once a variant is reviewed and saved in VSClinical, it will be stored in the appropriate catalog. A user may also create assessment catalogs without going through VSClinical. We have a wealth of information on our website regarding this topic, such as this very informative blog and our blog page which users can browse.
Using Assessment Catalogs
The default system within VSClinical automatically stores variants in assessment catalogs for tracking and information retrieval. Whenever the same variant is encountered, we will retrieve the interpretation and display the last edited date and the author of the interpretation.
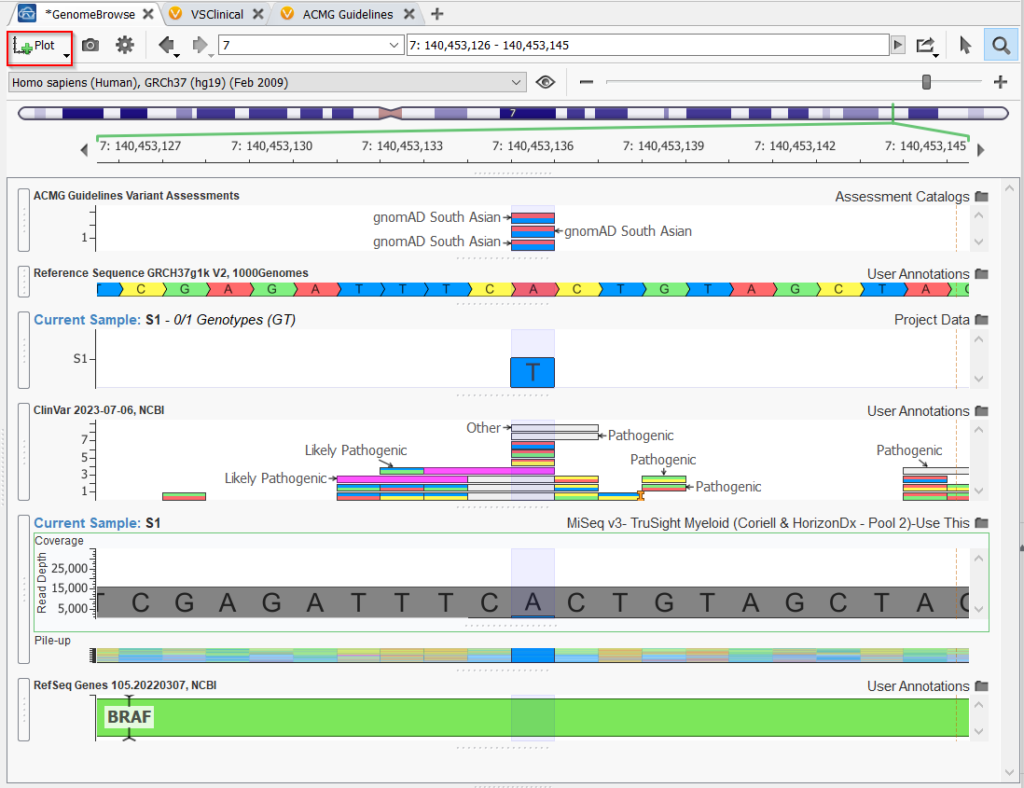
Two other very useful ways to use the assessment catalogs are to plot them in Genome Browse (Figure 4) or add them to the variant table as an annotation (Figure 5). The assessment catalogs are added to the user’s Local folder, and so can be accessed by going to Plot>Plot new source in Genome Browse or Add>Variant annotation in VarSeq.
We also have resources on using assessment catalogs for CNV frequency and for keeping track of variants that are sequencing artifacts. If you have any questions about creating or using assessment catalogs, or tracking variant interpretations or artifacts, please reach out to our support team at [email protected]. Thanks for reading!