Recently I gave a presentation on bioinformatic filtering: the process of using quality scores, annotation databases, and functional prediction scores to intelligently and quickly reduce your variant search space.
In this webcast, I mention that filtering is something we have been doing for a long time, and that there are some great examples that use exome sequencing data along with common variant databases in papers dating back to 2009. Given that we can now sequence cohorts to our heart’s content (well, at least our budget’s content), producing larger and larger repositories of data to support our research endeavors, bioinformatic filtering is a methodology that will remain critical to sequencing analysis workflows for a long time to come.
At the end of the day, we are really good at producing data, but there is great opportunity in both improving the quality of the data, and in using advanced techniques to help make sense of the petabytes of gigabases surging from sequencing machines all over the world.
This is a high level presentation regarding a very powerful methodology, and there is definitely room for discussion. Please feel free to post questions and comments.
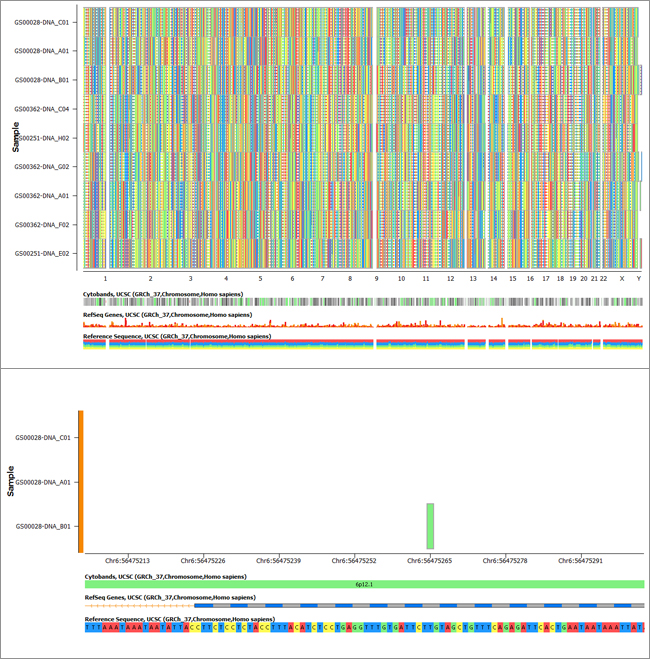
Two variant maps (used to visualize sequencing variants) showing the full whole exome data from 9 samples (top) and the result of the use of filtering tools to zero in on one family where the child has a de novo variant (bottom).