Recently, Golden Helix attended the ASHG 2025 conference, where we had the opportunity to showcase our partnership with Genomenon. This collaboration took center stage at both our booth and Genomenon’s CoLab session, Automating Genomic Workflows: Cutting Interpretation Time, Accelerating Turnaround, and Increasing Diagnostic Yield. Our discussions centered on a problem that every clinical lab faces as variant volumes continue to rise – manual interpretation doesn’t scale.
Scalable interpretation depends on pairing automation that labs can actually adopt with curated evidence that strengthens each decision. What stood out at the conference was the pressing need across the field. Our team enjoyed meaningful conversations with conference attendees, including many from labs eager to automate and scale their genomic interpretation in-house. Our enterprise platform, VSWarehouse 3, meets this exact need as an end-to-end software solution capable of generating reportable results from sequencing data, integrating comprehensive variant annotation sources to boost clinical diagnostic yield. At our booth, we demonstrated how these capabilities integrate seamlessly with the rest of the Golden Helix software suite, from VarSeq to VSPipeline.

What we showcased at ASHG reflects our long-standing broader mission to streamline variant interpretation and reporting, building on the automation and scalability pioneered in VarSeq and VSPipeline. In VSWarehouse, we’ve expanded on these features with a GUI to configure, run, and scale automated workflows, alongside full CLI orchestration. Furthermore, dynamic compute agents enable these workflows to run flexibly across local (on-premises) or cloud infrastructure. Together, these capabilities position our enterprise software suite as a robust, scalable environment for high-throughput clinical variant interpretation.
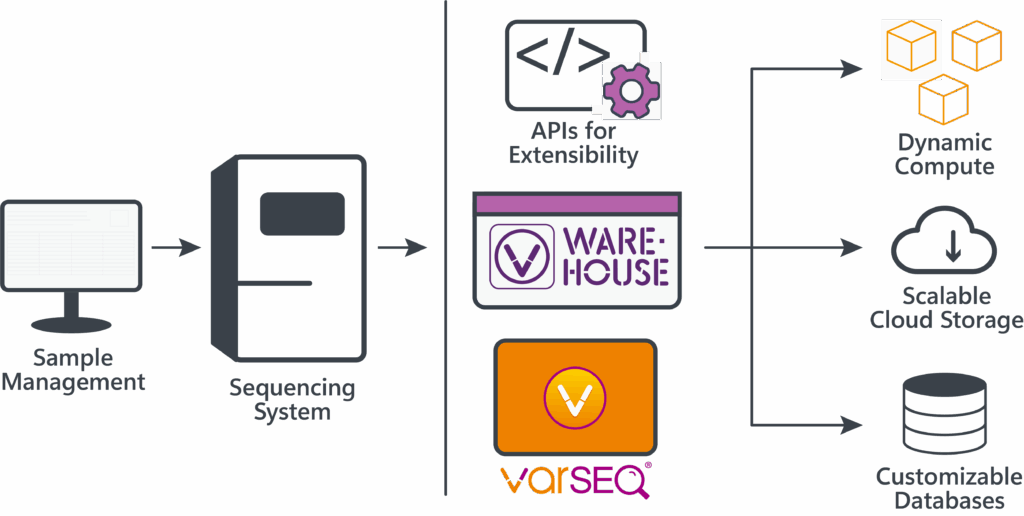
In large-scale whole-genome interpretation, success is measured not only by throughput but by the accuracy and reliability of the results. This is why our software suite integrates a broad range of curated databases and algorithms to enhance variant annotation, filtering, and classification. Clinical variant interpretation improves as annotation sources become more comprehensive and diverse, thereby reducing the risk of false negatives. That’s where Genomenon’s comprehensive databases are shining a light on variants that would otherwise go unnoticed, such as deep intronic variants with functional consequences. At the booth and during the CoLab presentation, we walked through examples showing how Genomenon’s curated evidence can change the outcome of an interpretation in VarSeq and VSClinical. By integrating Genomenon’s curated resources directly into VarSeq, workflows can more effectively identify clinically relevant germline and somatic variants.
ASHG 2025 underscored the importance of scalable, accurate interpretation in clinical genomics—a focus we share with our partners at Genomenon. To learn more, check out our recent Webcast about how Golden Helix and Genomenon are partnering to bring curated databases from Genomenon Mastermind and Cancer Knowledgebase into VarSeq and VSWarehouse workflows.