The recent release of VarSeq 2.6.0 was filled with so many customer-requested features (for example, our long-awaited PGx workflow!) that some of our other new features have not yet had their time in the spotlight. For this blog, we are thrilled to announce that with the release of VarSeq 2.6.0, we have made WGS CNV calling more user-friendly than ever with several new upgrades!
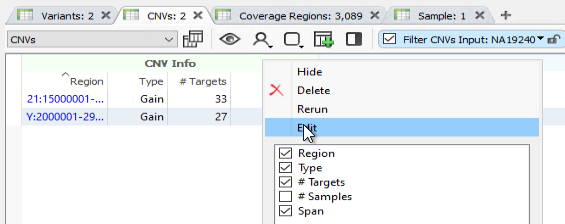
First off, our Binned CNV caller now has an edit button (Figure 1)! This feature will allow WGS workflows to easily access the Binned CNV caller settings for editing without having to delete and rerun the whole algorithm from scratch. This simple change will help streamline the workflow development process for our customers who work in the WGS space.
In addition to the Binned CNV Edit button, we have also increased the accuracy of our caller for those large CNV events. By merging adjacent CNVs into a single event when there are no deviating targets between CNV calls, we can more accurately identify large deletion events. You can see this clearly in Figure 2, where part A shows the presence of multiple Het Deletions across a gene. Instead of calling this as so many smaller CNV events, Figure 2B shows one much larger Het Deletion across the entire gene. For more information about the accuracy of our CNV caller, please check out this webcast on our CNV caller with the Twist Exome 2.0 Plus Comprehensive Exome Spike-in capture panel.
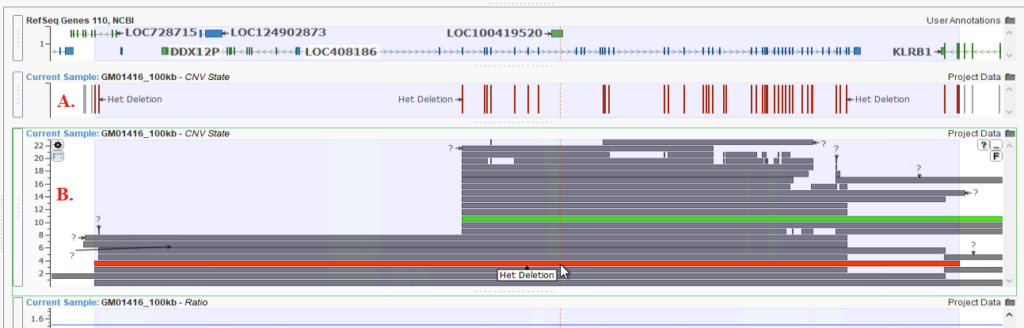
B: The Het Deletions being called correctly as one larger CNV event.
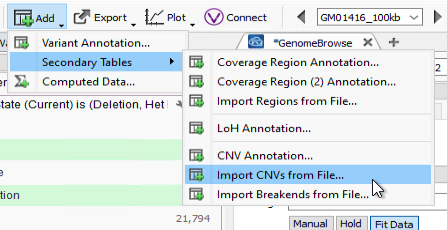
We also have good news for those customers who are not using our CNV caller but are importing externally called CNVs. As long as externally called CNVs are formatted into TSV files, we are now able to import and analyze that data in both VarSeq and VSClinical. To try this out, go to Add > Secondary Tables, and then Import CNVs from File to bring in those externally called CNVs (Figure 3).
Last but not least, on our CNV-centric upgrades to VarSeq 2.6.0, our LOH caller has been significantly improved in terms of speed! We have done this by optimizing the way this algorithm reads the VAF and GQ values in each chromosome for all samples in a project at once.
Through these improvements, we hope to streamline and optimize our customers’ workflow experience. If you have any questions about how any of these upgrades can benefit your workflows, please contact our FAS team at [email protected]. We will be happy to chat about your unique use case needs.