ClinVar is one of our most used annotations sources for a variety of workflows. It is also the public annotation source that is updated most frequently of all the sources currently supported in VarSeq. ClinVar provides new versions of their database once a month in several formats (XML, VCF, TXT). We use custom Python scripts to convert the provided VCF and text data into annotation sources that can easily be used in any of the Golden Helix products.
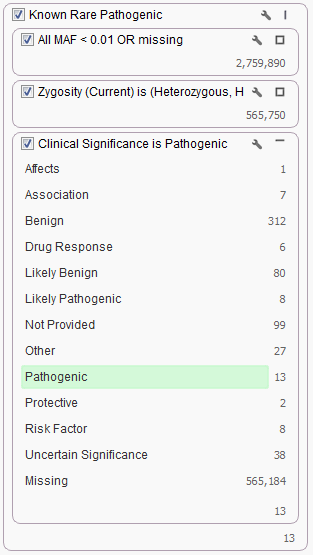
ClinVar can be used for annotation purposes in addition to filtering your data. The most common filter choice for this source is using the Clinical Significance field. For example, in our Exome Trio Analysis Template, the Known Rare Pathogenic filter chain looks for only those rare variants classified as Pathogenic by ClinVar.
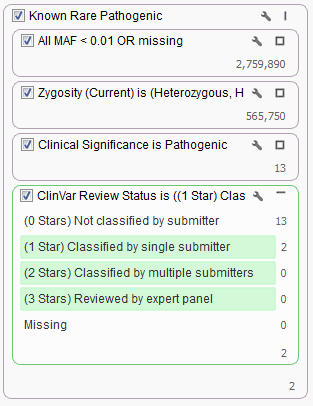
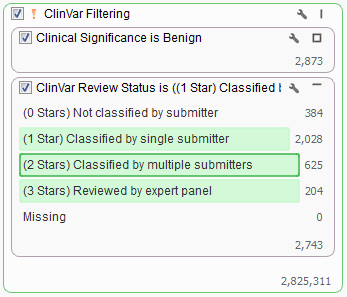
What if you want to also incorporate a second field from ClinVar into your filter chain? For example, what if you only want to look at those Pathogenic variants that have been classified by at least one submitter? You can do so by adding a filter card using the ClinVar Review Status field and keep all but the “Missing” and “Not classified by a submitter” categories.
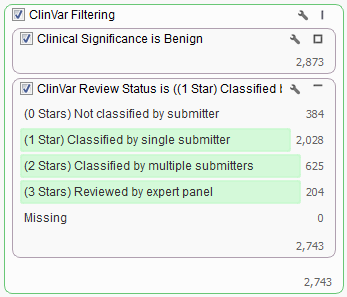
Filtering workflows can get even more complex when using ClinVar. For example, say you would like to exclude all variants with “Benign” status and at least a 1 star classification whereas variants with status other than “Benign” should remain in the set regardless of their star classification.
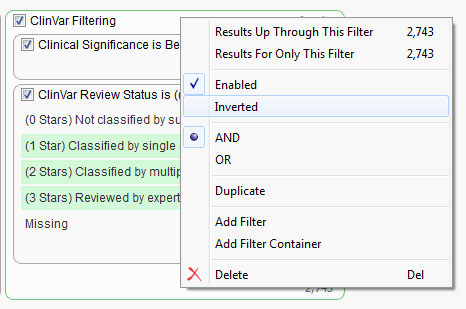
When looking to filter certain variants it is sometimes easiest to filter down to those variants you want removed and then invert your filter selection instead of directly selecting those you want to keep. In the case above we would first want to find all “Benign” variants based on the Clinical Significance field then add in the ClinVar Review Status and select all categories other than “Not classified by submitter” or “Missing”.
In this dataset there are over 2,700 variants that we want to be filtered from our list. To facilitate this both cards were added to the same filter container “ClinVar Filtering”. You can create a filter container by right clicking in any empty space on the Filter View and adding a Filter Container.
Once the container is built and the filter cards have been added we now want to invert the selection which will remove the identified variants from our workflow. Just right-click on the name of the container and select “Inverted”.
You should then be left with something like the following as the final results.
Please let us know at [email protected] if you would like assistance building your custom filter chain or if you have any questions about your VarSeq workflow.