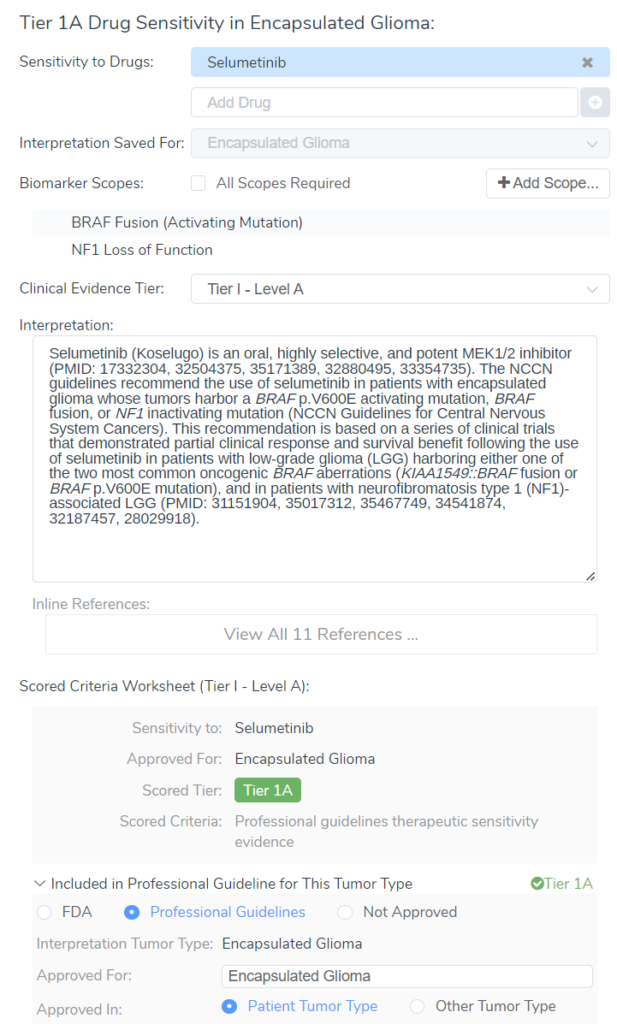
I am very excited to announce that a new version of Golden Helix CancerKB has been released! This new version of CancerKB comes with some exciting upgrades bolstering the gene interpretations and cancer gene evidence. Before I jump into the details of the new information that is now in CancerKB, I want to give everyone a quick reminder of what the CancerKB database has to offer. CancerKB is a collection of professionally curated cancer interpretations for genes and biomarkers, paired with clinical evidence for drug sensitivity and resistance, and diagnostic and prognostic evidence.
CancerKB is methodically curated utilizing well-established cancer databases, biomarker frequency resources, literature review, and application of FDA, EMA, NCCN and other professional guidelines for therapeutic information. CancerKB is unique from other cancer biomarker databases because it actually provides written interpretations that are consistently formatted and ready for clinical reports as opposed to other sources that simply provide some data, but this information is not formulated into something readable or transferable to patients or clinicians.
In CancerKB 3.0, using the same methodical curation process described above, cancer gene curation was enhanced, leading to the release of the Golden Helix CancerKB Gene source. This source simply itemizes the information that is in the written gene interpretations so these data can more easily be incorporated into your workflow. This new source can be used in VarSeq projects for filtering and has been integrated for use within VSClinical. Golden Helix CancerKB Genes captures:
- The gene’s role in cancer is the gene an oncogene, tumor suppressor gene, or both.
- The gene’s cancer hallmarks, according to the updated cancer hallmark’s publication.
- Relevant somatic tumor types, the most prevalent tumor types associated with the gene
- Relevant germline cancer predisposition tumor types, if applicable
- The most common cancer-causing mutation types for the gene, such as fusion, missense, splicing, or copy number changes
In addition, the gene-focused curation resulted in the curation of ~500 more cancer genes, meaning CancerKB now describes roughly 1,000 genes and their relevance in cancer. The genes described cover not only the most well-known cancer genes but also several emerging cancer genes.
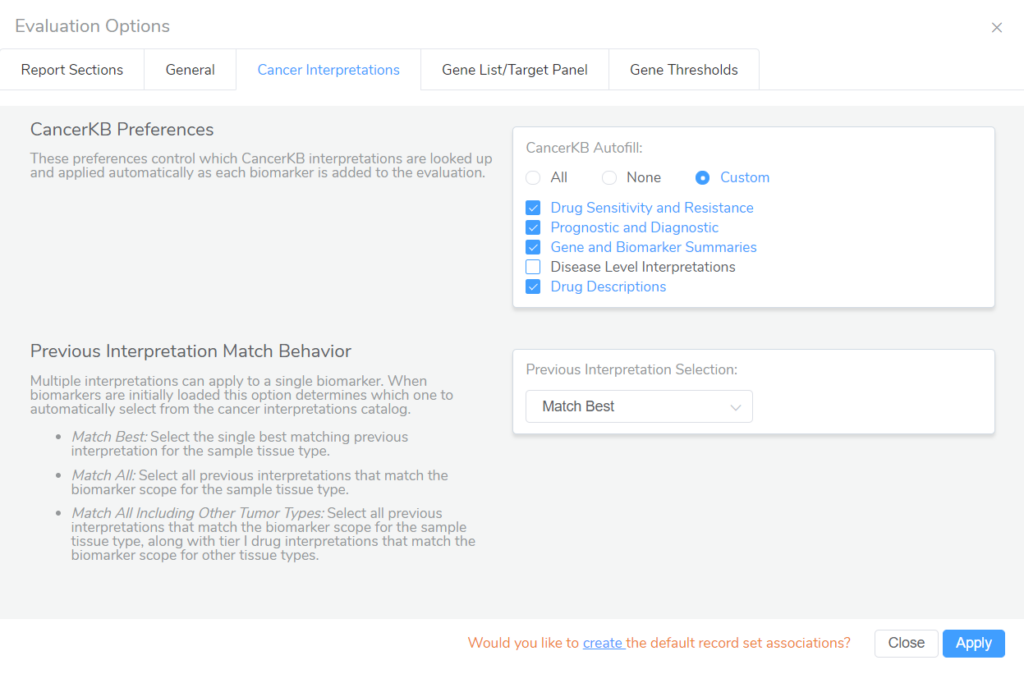
The last thing I want to mention pertaining to CancerKB is that the recent VarSeq 2.5.0 release came with some added control over which CancerKB interpretations will autofill into VSClinical evaluations. In the Evaluation Options for the AMP Workflow, the Cancer Interpretations tab allows users to decide which CancerKB interpretations will fill in to patient evaluations, along with setting the preference for previous interpretation matching behavior.
Golden Helix CancerKB is updated monthly to keep interpretation fresh and accurate and contains interpretations for the latest biomarkers and drugs as new therapies are approved. We are always working on curating new material that continually makes CancerKB stand out as a must-have resource in your cancer workflows. Stay tuned for what is coming up next for CancerKB!