There are several contextual factors to consider when analyzing genomic data for NGS analyses. A variant may have divergent impacts depending on which transcript of the gene is being considered, or the impact of a variant could be weighed more or less heavily depending on the disease context. Evidently, the user’s gene preferences are very important, but not all software tools make it easy to take this into account. VarSeq is one of those software tools that gives users control over their gene preferences, enabling them to fine-tune the context of their analysis. This blog will discuss a new VSClinical feature, the Gene Mutations and Preferences tab in VSClinical ACMG, making managing user gene preferences even easier.
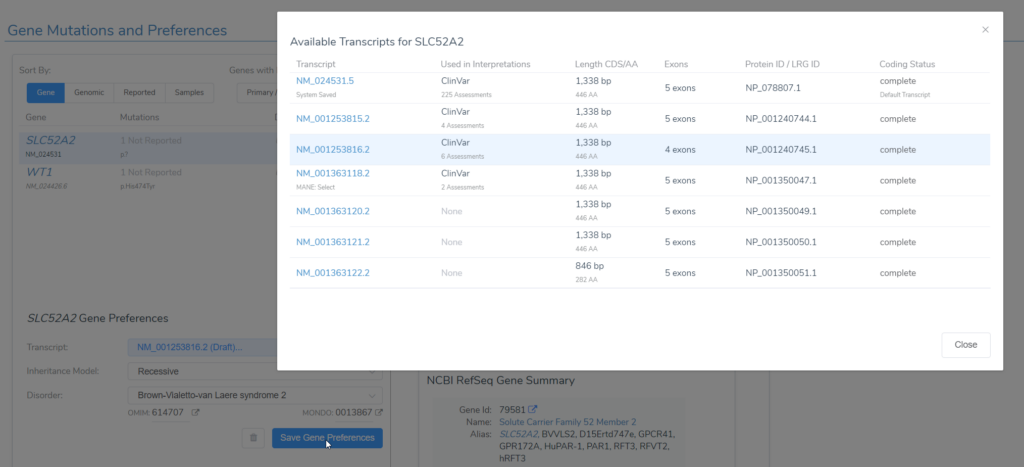
The software system preference for default transcript selection for VarSeq annotations and variant interpretation in VSClinical is the clinically relevant transcript, which is usually the MANE select transcript. Please see this excellent blog on how VarSeq arrives at the clinically relevant transcript designation for each gene and how we provide both combined and clinically relevant functional impact annotations. In some cases, the system gene preferences override the selection of a clinically relevant transcript as the default, for example, when there is a transcript besides the “clinically relevant” one that is being used predominantly in the public domain for a particular gene. Regarding gene-disease associations, we typically default to the most significant but acknowledge that any given user may be operating in a different context. The automation of transcript selection and gene-disease association is great, but sometimes, users need to go behind the curtain and customize their workflow. VarSeq enables the user to do just that, and our new Gene Mutations and Preferences tab in VSClinical allows users to control their gene preferences from the GUI, precluding the requirement for any special scripting skills.
The simplest approach to control the user-preferred transcripts would be through the RefSeq Genes track annotation, where you can add preferred transcripts in bulk. This only allows users to modify preferred transcripts but not gene-disease associations and inheritance modes for particular genes. To do all the above, a user can also edit the user gene preferences file, as explained in this blog, but this involves scripting and thus may be inaccessible to some users. Now, the Gene Mutations and Preferences tab in VSClinical ACMG allows the user to easily and quickly, on a gene-by-gene basis, edit preferred transcripts, change inheritance model, and change the disorder or disease associated with a gene with a few clicks of a button. To reset the defaults, simply delete the gene preference entry by clicking on the trash can next to the Save Gene Preferences button.
In keeping with our culture of giving users great automation capabilities yet allowing them to retain much control over their analyses in our software, this Gene Mutations and Preferences feature has made it super easy to manage user gene preferences. Please let us know at [email protected] if you have any questions or comments about this topic.