Curated databases are a real time saver when compiling published evidence to support your variant evaluations and classifications. Leveraging the curated databases at your fingertips in our VSClinical variant interpretation hub is even more efficient. Not only does VSClinical provides users with automated variant classification for germline variants according to the ACMG guidelines and somatic variants according to the AMP guidelines, but we also give users immediate access to the curated databases and literature sources that are relevant for their evaluation. This blog is meant to highlight the curated databases such as ClinVar, dbSNP, PMKB, COSMIC, and others that users have access to and describe how to leverage them in VSClinical.
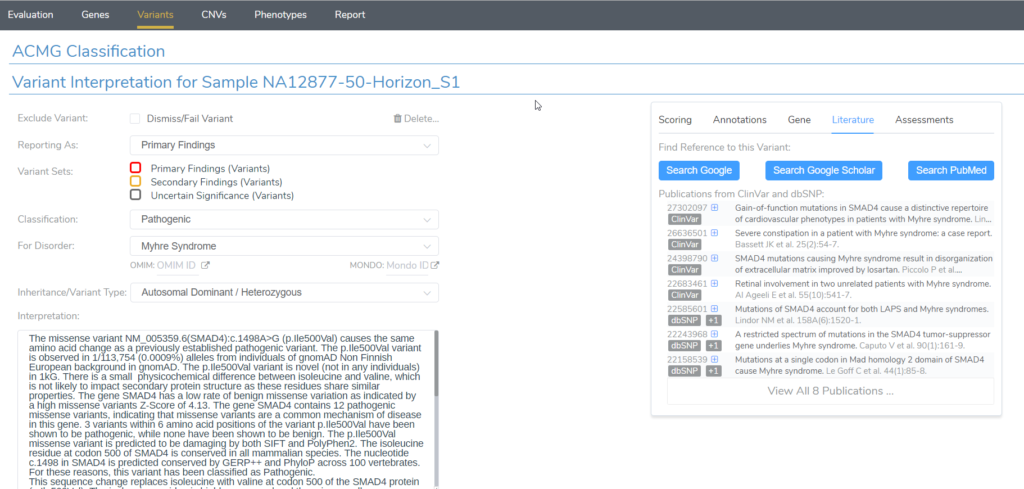
When evaluating a germline variant in VSClinical ACMG, we often focus on the Scoring tab in the Variant Interpretation section of the Variants window. Just switching over to the Literature tab here uncovers a wealth of variant-specific literature references that users can easily explore from the ClinVar and dbSNP curated databases. Users can easily copy any of the citations by clicking on the blue plus icon next to each record.
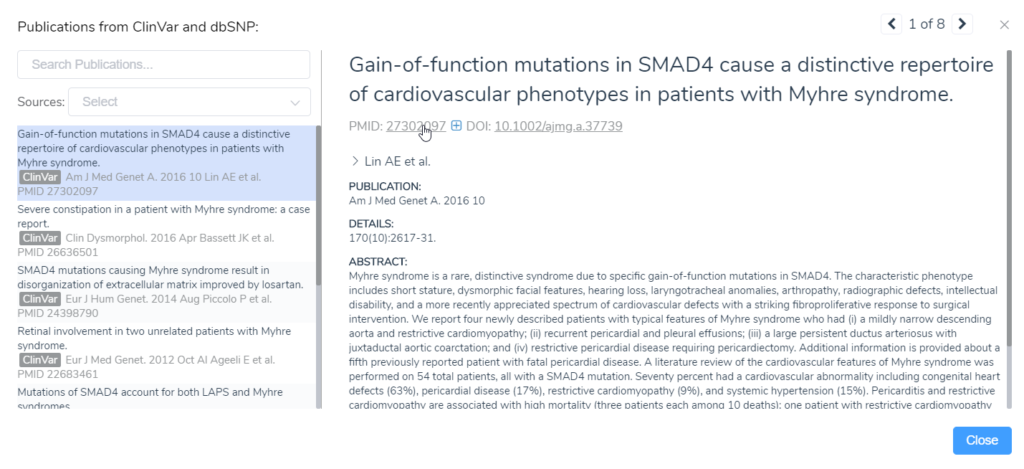
Clicking on any record opens up a more detailed window. Here you can quickly overview each publication by reading the abstract directly in VSClinical. You will also find links to the PubMed and DOI databases.
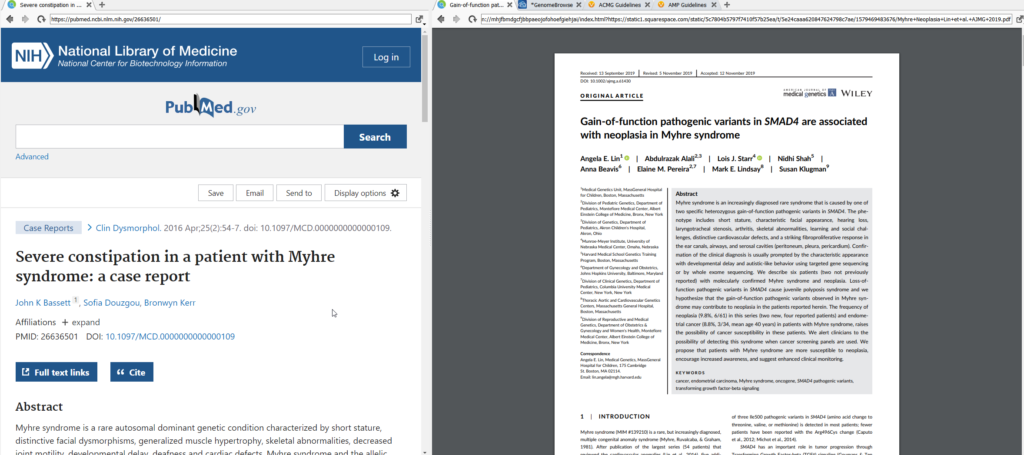
Clicking on the PubMed link will open up that site in a web browser, while the DOI link will pull up all the specific Google searches for that record, including any available PDFs.
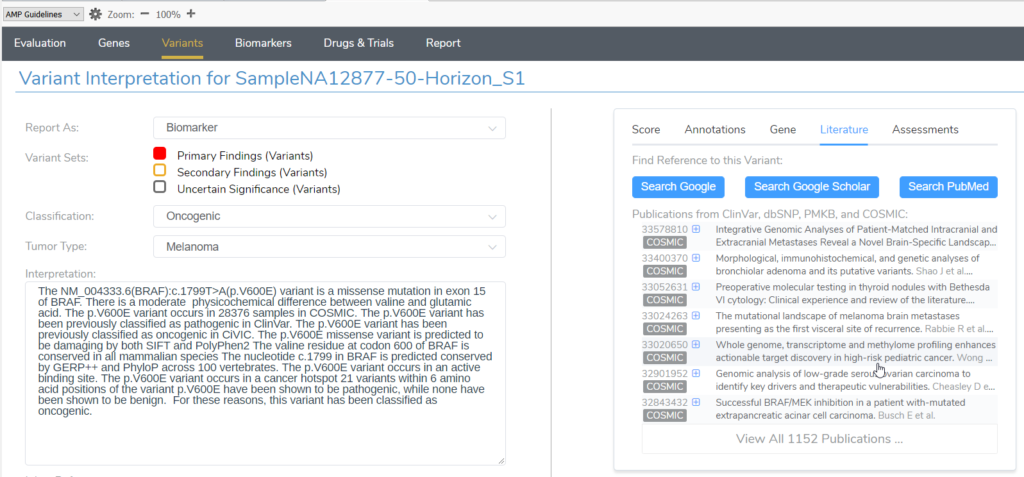
Similarly, users doing somatic variant evaluation in VSClinical AMP can access even more curated databases. The selection here is expanded to include publications from PMKB and COSMIC in addition to ClinVar and dbSNP.
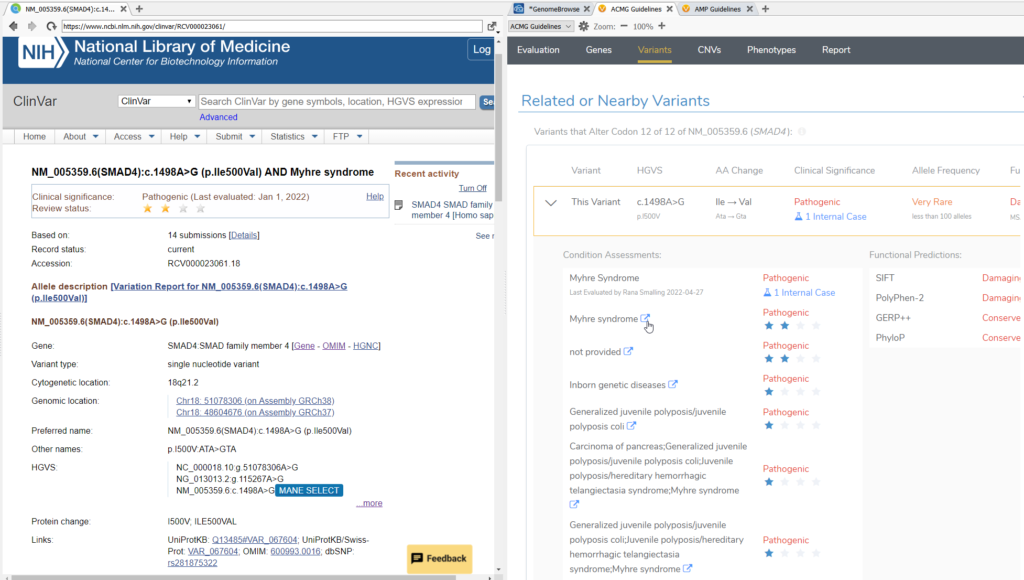
The Related and Nearby Variants section of the VSClinical ACMG Variants window will allow you to explore the ClinVar curated database for submissions and publications about the variant under evaluation or other nearby variants in relation to specific disorders. In VSClinical AMP, the same function is achieved under the Relevant Clinical Assessments section.
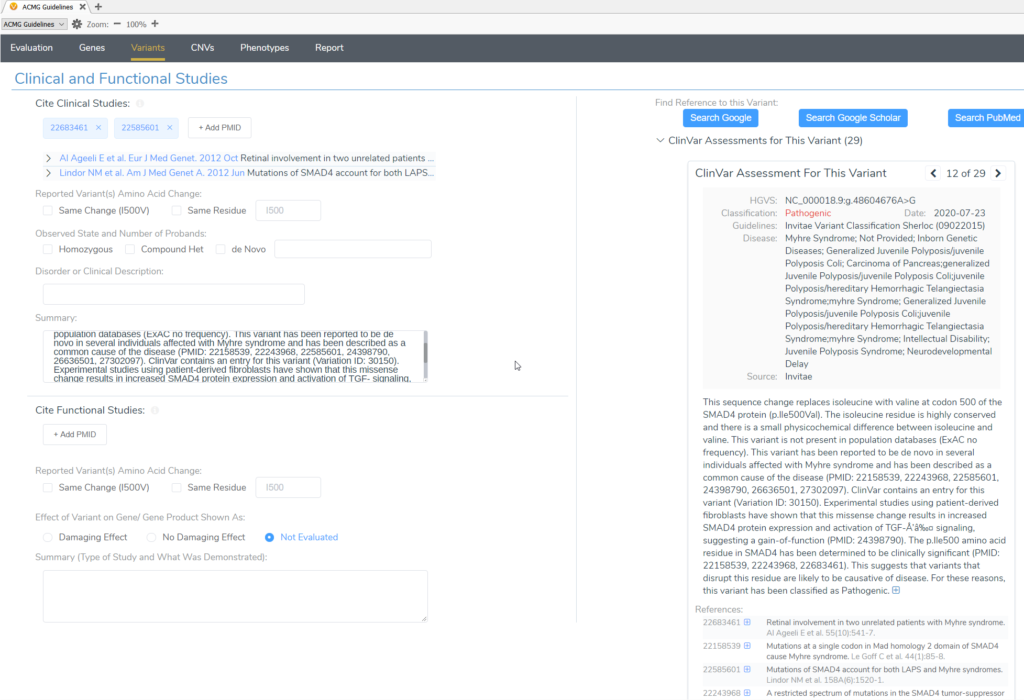
The final resource for curated databases in the Variants window of VSClinical ACMG is the Clinical and Functional Studies section. Here, you can find variant-specific and residue-specific ClinVar submissions with publication references, if available.
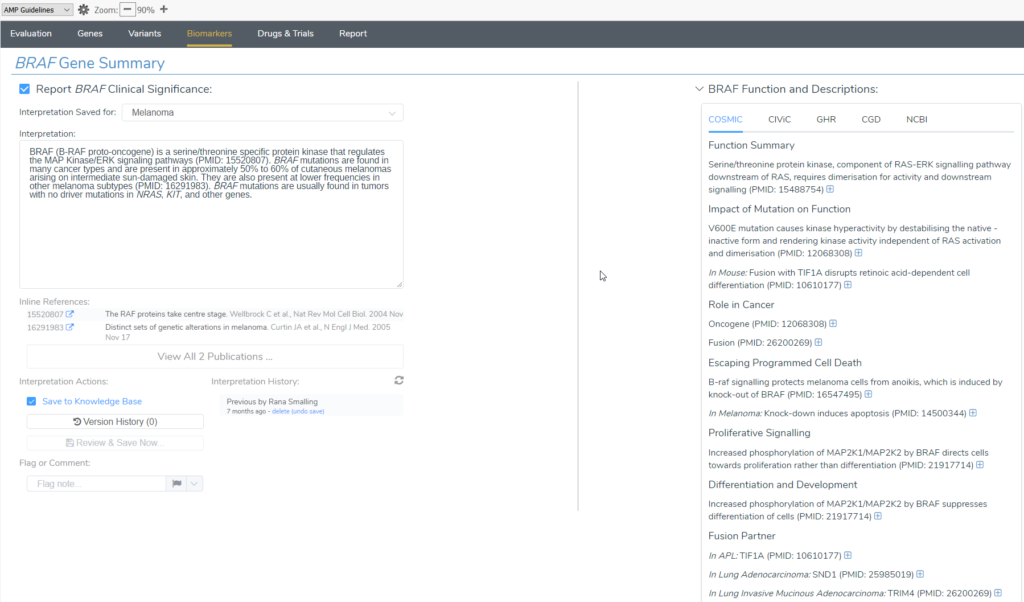
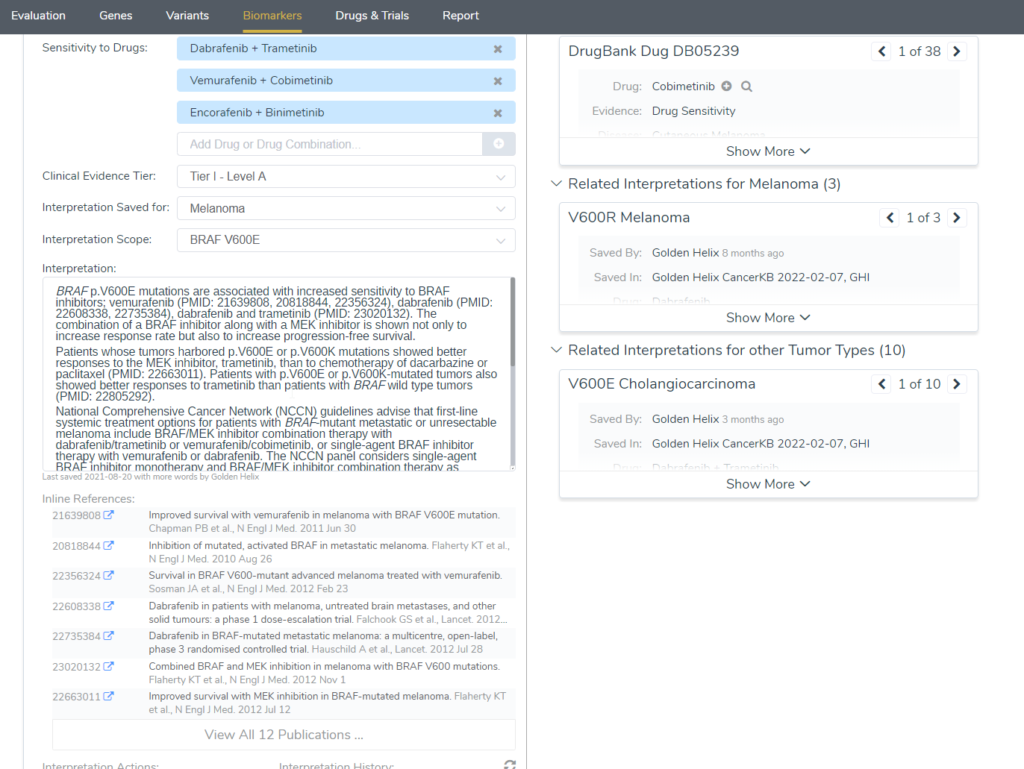
VSClinical AMP offers users evaluating somatic variants a dedicated window for Biomarkers analysis. Typically users quickly home in on the available treatment options listed in the Clinical Evidence section. However, there are copious amounts of literature here to be mined as well. Notably, this is where users can access our very own CancerKB, a curated database of interpretations of biomarkers for a variety of cancer types.
In the Gene Summary section users can leverage the automated CancerKB interpretation of the specific variant and will be able to explore records available from the COSMIC, CIViC, GHR, CGD, and NCBI curated databases. These references and notes may be variant or gene-specific and include literature about related fusions.
Below these databases we have a dedicated CancerKB tab with specially curated interpretations about the specific variant in other tumor types with literature references.
CancerKB interpretations with references are present throughout every section of the Biomarkers tab including the Alteration Frequency and Outcomes, Biomarker Summary, and Clinical Evidence sections. The Clinical Evidence sections even includes interpretations of the drug treatment options available and the associated references.
VSClinical automated variant evaluation and classification are simultaneously seamless and transparent. Any user can quickly review their variant evaluation and move on to the clinical report or spend as much time as required to delve into the curated databases and literature-based evidence that were used to arrive at the final classifications.
I hope you found the contents of this article useful. Should you have any questions, comments, or concerns about this blog, please reach out to us at [email protected]. Thank you for reading!