Golden Helix’ SNP & Variation Suite (SVS) has a Regression Module to enable researchers with varying degrees of statistical knowledge to interrogate their data using regression models to account for potential confounding effects of covariates and interaction terms. While these tools are labeled “basic”, they can be difficult to use and results hard to interpret for those who have only had a course or two in regression analysis; even more seasoned researchers have a hard time determining how to set up a model.
This blog post is designed to be a thorough introduction and provide more details on how to set up linear regression models than what is currently provided in either the SVS Manual or our tutorials. This post will explain:
- How to do a simple regression model that tests every genetic marker against one dependent variable.
- How to adjust for covariates and run a full versus reduced model.
- How to look at SNPs in a range to see if adjacent markers have a combined effect on the dependent variable.
- A few common FAQs regarding running regression.
Linear versus Logistic Regression
Just as a refresher, a linear regression model takes a quantitative dependent variable (either integer or real-valued) and fits a “line” to the data. A logistic regression model takes a binary (0, 1) dependent variable and fits a “line” to the data after a logistic transformation. The models are set up the same way but some of the interpretation is different. See the Regression with Covariates tutorial for information specific to logistic regression models and output. This post will focus solely on linear regression.
A note before we get started
Golden Helix SVS requires that all regressors be numerically encoded. A regressor is the primary independent variable or variables used for either the “Regress on each numeric column” or “Moving window regression” options. Additional covariates added to the model through the full model covariate or reduced model covariates box can be numeric or categorical variables.
This means that if your genetic markers are SNPs encoded as genotypes, you will first need to recode these genotypes into numeric values. In SVS, go to Edit > Recode > Recode Genotypes and select either an Additive, Dominant or Recessive model. If your genetic markers contain Log Ratio data, no recoding is necessary.
Running every genetic marker against a dependent variable
The best regression model to run first is one dependent variable against all SNPs individually. In this case, the hypothesis is that there is no relationship between a particular genetic marker and the dependent variable. If you have N genetic markers, the Regression Module will run N regression models. You only have to set up the Regression window once and all of the models will be run and the results compiled into a spreadsheet.
Regression Model:
Dependent Variable = Bo + B1*Genetic Marker
In SVS this looks like:
 Figure 1: Simple Linear Regression Model; no covariates
Figure 1: Simple Linear Regression Model; no covariates
Results Look Like:
 Figure 2: Simple Linear Regression Results; sorted ascending on Full-Model P-Value
Figure 2: Simple Linear Regression Results; sorted ascending on Full-Model P-Value
Meaning of Columns (i.e. Where do I look to determine significant models?)
| Column | Meaning |
| Full-Model P-Value: | Probability of observing a test statistic as extreme as or more extreme than the one observed, given that the underlying assumption is that the coefficient being tested is zero. In other words, look at this value when trying to determine statistically significant regression models. |
| Mean Y: | Mean value for the dependent variable. |
| Y-Intercept: | Intercept of the regression model. |
| Residual SE: | Standard error of the residuals for the regression model. |
| Slope: | Coefficient or estimated values attached to the regressor (SNP). |
| Slope SE: | Standard error of the coefficient. Obviously the estimated coefficient is not the “true” value, therefore, an associated error is incurred for the regressor (SNP) and is reported in this column. |
| F: | The F statistic for the regression model, used to calculate the Full Model P-Value. |
| Sample Size: | Number of samples used in the regression model. If the dependent variable was missing for a sample, the sample would be dropped from the analysis. |
Adjusting for confounding variables and then determining effect
A more complex regression model is to determine the effect of genetic variables after adjusting for confounding variables. The question answered by this model is how much contribution to a regression model does the genetic variable have after you remove any confounding effects that age or gender or both might have on the response. Some response or dependent variables have known associations with age and/or gender and so it is important to remove these effects before trying to determine if there are any genetic markers associated with the dependent variable.
This is one of the more confusing models to set up in SVS. While the parameters themselves are pretty straight forward, many of our customers find themselves at a loss as to what to do until they have set these models up a few times. Let’s first consider what we want the model to look like.
- We want to determine if there is any relationship between the covariates and the dependent variable and if there is, we want to remove this effect.
- After that is done, we then want to add in the genetic marker and see if the marker adds significantly to the regression model or not.
To accomplish this, we will need two regression models. We call the model with the covariates that we want to treat as confounding variables the “reduced model”. We don’t care about these covariates other than to remove their effects from the model. We call the model with the genetic marker or additional regressors in addition to the covariates, the “full model”. To get the significance of the genetic marker after removing the confounding variables, we want to perform a “full versus reduced model”.
Reduced Model (covariates are both age and gender):
Dependent Variable = B0 + B1*Age + B2*Gender
Full Model (regressor is SNP1):
Dependent Variable = B0 + B1*Age + B2*Gender + B3*SNP1
In SVS this looks like:
 Figure 3: Linear Regression – Full Vs Reduced Model Correcting for Age and Gender
Figure 3: Linear Regression – Full Vs Reduced Model Correcting for Age and Gender
Results Look Like:
 Figure 4: Linear Regression Results – Full Vs Reduced Model Correcting for Age and Gender; sorted ascending on FvR Model P-Value
Figure 4: Linear Regression Results – Full Vs Reduced Model Correcting for Age and Gender; sorted ascending on FvR Model P-Value
Meaning of Columns (i.e. Where do I look to determine significant models?)
| Column | Meaning |
| FvR Model P-Value: | Probability of observing a test statistic as extreme as or more extreme than the one observed , given that the underlying assumption is that the coefficient being tested is zero. In other words, look at this value when trying to determine if the regressor adds significantly to the model after correcting for the covariates (age and gender in this case). |
| Full Model P-Value: | Contains the p-value for the regression model that contains all of the covariates and regressors. |
| F Full vs Reduced Model: | The F statistic used in determining the p-value for the full versus reduced model p-value. |
The other columns contain the same information as in the first “simple” linear regression model. See above for the details on those columns.
Adjacent markers having a combined effect
To look at SNPs in a range to see if adjacent markers have a combined effect on the dependent variable, you need to do a moving window regression. You can either specify number of columns to consider at once or a dynamic window that is determined by distance in base pairs. To perform dynamic moving window regression, it is required that you have a marker map applied to your spreadsheet. When performing this regression you can also correct for covariates if you wish.
Say that you want consider a window of 10,000 kb with a maximum of 10 markers in the window.
In SVS this looks like:

Figure 5: Dynamic Moving Window Linear Regression
Running this model internally produced p-values, the lowest of which were around 0.0001. To get information on which markers were used in the regression models with the smallest p-values, adjust a parameter on the “Output Parameters” Tab to get detailed results and run the regression analysis again.

Results Look Like:
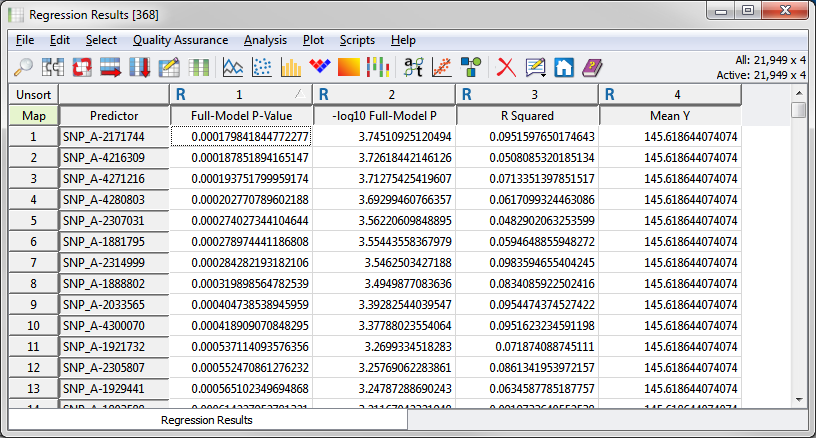 Figure 7: Dynamic Moving Window Linear Regression Results; sorted ascending on Full-Model P-Value
Figure 7: Dynamic Moving Window Linear Regression Results; sorted ascending on Full-Model P-Value
 Figure 8: Dynamic Moving Window Linear Regression – Detailed Output for Models with Full Model P < 0.0005
Figure 8: Dynamic Moving Window Linear Regression – Detailed Output for Models with Full Model P < 0.0005
Frequently Asked Questions:
Q. I am trying to run a regression model on all of my numerically encoded genotype data, and I keep getting an error that the regression failed. Why?
A. You are most likely specifying too many covariates for the model; this could be because you are not setting up the model correctly.
If you want to regress each numerically encoded genotype data against the response one at a time then you need to select the “Regress on each of the N numeric columns” option under “Selection Parameters”. If you want a model that includes multiple genetic markers and considers the interaction between all of the markers and the response, SVS’ regression package is not appropriate for this analysis.
If you just have several covariates such as age, gender, blood pressure, treatment, smoker, etc. and you get this error, try a stepwise regression. Usually if you are over specifying the model with all of your covariates, forward selection is the better stepwise approach as it adds one covariate to the genetic marker at a time until the p-value for the full model exceeds the specified p-value threshold.
Q. In the detailed output there is a univariate fit column; what does this mean?
A. This is the p-value resulting from the comparision of a model with only the associated covariate and an intercept term to a model with the intercept only and no covariates.
Q. Can SVS perform more complicated regression models, such as handling random effects or conditional regression?
A. No. The regression module in SVS is only designed to cover the basic regression needs of most researchers. If more complex regression models are needed, we recommend you use a software package designed to handle these regression models and encourage you to then import your data back into SVS for visualization.
Q. Can you help me interpret my regression results? How do I know what models are significant and which SNPs should I investigate further?
A. Here at Golden Helix we will do everything we can to help you get your regression results and understand the output you obtain. However, determining significance and which markers are worthy of further study is a personal research question that is best answered by the team that understands the phenotypes, confounding variables, and study population.
If you do not have statisticians available to help set up the appropriate models for your regression questions and would like assistance to do so, our services department is available to help!
And as always, if you run into problems, don’t hesitate to contact the support team at Golden Helix. …And that’s my 2 SNPs.
I have a question, i am creating an econometric model using multiple regression. And my problem is that every independent variable I have, using P value seems to be insignificant to the model 🙁 what can i do to make one or more of my variables would be significant enough?
Hi, Im little confused about odds ratio in regression output table of FvR model (SVS v8), if genotypes was recoded for additive model with three values. Whati is the meaning of this odds ratio? What is the formula to get it? Thanks for your answer, in advance.