I’m a believer in the signal. Whole genomes and exomes have lots of signal. Man, is it cool to look at a pile-up and see a mutation as clear as day that you arrived at after filtering through hundreds of thousands or even millions of candidates.
When these signals sit right in the genomic “sweet spot” of mappable regions with high coverage, you don’t need fancy heuristics or statistics to tell you what the genotype is of the individual you’re looking at. In fact, it gives us the confidence to think that at the end of the day, we should be able to make accurate variant calls, and once done, throw away all these huge files of reads and their alignments, and qualities and alternate alignments and yadda yadda yadda (yes I’m talking BAM files).
But we can’t.
Thankfully, many variants of importance do fall in the genomic sweet spot, but there are others, also of potential importance, where the signal is confounded.
Confounded by what? It’s tempting to say the signal is confounded by noise. And in the analog world, a signal with too much noise is a lost cause. Despite what nearly every detective show in the world tells us, you can’t take a grainy image and say “enhance” and get a high-definition rendering of the suspect.
But thankfully, the world of genomics and Next Generation Sequencing is not an analog world. And there are very discrete and tractable reasons why some loci and types of variants cause us so many problems.
And for the most part, we understand these problems!
Let me enumerate the biggest ones:
- Variants being called individually and not in a haplotype aware manner.
- GC rich regions or regions that produce PCR or sequencing artifacts
- Regions of the genome with low mappability or high homology to other regions either large or small.
- Poor homology between the sample’s genome and the reference. For common reasons such as:
- Structural variants like inversions or translocations in the sample.
- Sites with tandem repeats or mobile element insertions.
- Sites where the reference sequence is unable to capture the allelic diversity of the population and the sample (like the MHC region).
Not to mention, when looking at a sampling space so large, very rare events from a statistical perspective will occur. So if you have a 30x genome, you should expect a heterozygote on average to be covered by 15 reads to support the mutant allele. However, there will be loci where a heterozygote legitimately exists but it gets sampled only once or twice. (The chances work out to expecting to see this 1.74 times when calling 4 million variants). Far from a convincing number to distinguish it from a sequencing error.
These issues are not new, and we have even relegated their effect to a minority of variants. In fact, it seems like for years the assumption has been that we are just a small incremental step away from having variant calling licked.
I remember reading a blog post in mid-2011, by the Chief Science Officer at Complete Genomics, pitching that we should be throwing away our read alignment data in most cases. Variant files with quality scores should be good enough. Otherwise go resequence the sample!
But in reality – and I watch this stuff closely – over the last three years we have made very little progress towards handling these systemic issues.
And that’s pretty astounding.
One Cell Line to Rule Them All
Since the products we create at Golden Helix for doing variant analysis and interpretation start after variants have been called, I have become intimately aware of the state of data our users receive from their upstream sequencing provider.
Recently, I decided to do a more thorough comparison of the same samples’ variant calls from a representative set of three sources: Complete Genomes, the Illumina Genome Network, and a typical Core Lab’s pipeline (hosted by the 1000 Genomes folks at the Broad Institute).
Luckily for my purposes, a specific trio from the CEPH extended pedigree (Utah pedigree 1463) has become the de facto standard for benchmarking sequencing platforms and secondary analysis pipelines.
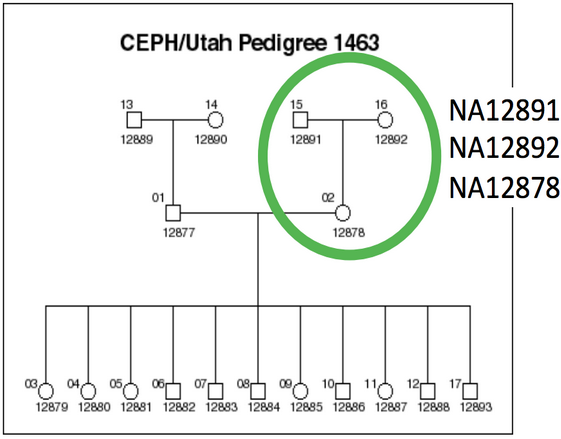
The trio used for comparing different service providers highlighted in the publicly available and highly sequenced Pedigree 1463. (Credit: Steve Lincoln, Complete Genomics)
The NA12878 daughter and parents trio was included in the HapMap project, the 1000 Genomes Project and is available to any lab from Coriell as a cell line (for ~$85 a sample).
In fact, NA12878 is easily the most sequenced genome in the world!
Complete Genomics’ data is available on their website, Illumina provided me with a hard drive containing standard IGN deliverables for the trio, and with a little digging, I found a comparable genome at 60x coverage produced by a pipeline that matches the Broad’s GATK best practices (one that many core labs model their deliverables on). The latter I refer to as the “BWA/GATK” pipeline and will reflect many pipelines in production to produce both research and clinical use exomes and genomes.
So with SVS as our workhorse, it was no problem at all to import all three of these source’s variants and do a little comparison.
So here is the big picture:
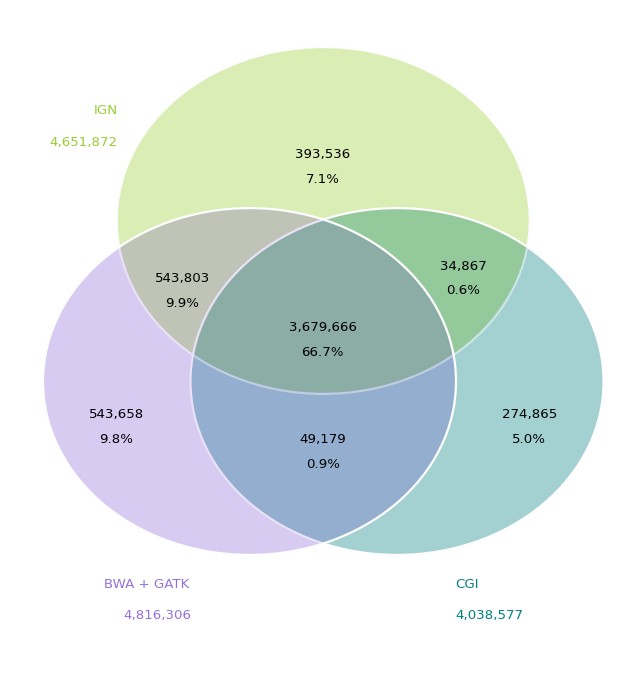
Looking at the concordance of the full variant sets for NA12878 between Illumina Genome Network, Complete Genomics, and a BWA/GATK pipeline.
Note that both IGN and the BWA/GATK pipeline are based on HiSeq 2000 which produced 100bp paired-end reads. So they use the same sequencing technology, but significantly different alignment and variant calling techniques. IGN uses the ELAND aligner and CASAVA variant calling, versus BWA for alignment and then various steps to clean those up (de-duplication, quality score recalibration and local indel re-alignments) before calling variants with GATK.
The Illumina based sources do agree on another 10% of the variants not also called by Complete Genomics. Given that Complete Genomics uses an entirely different sequencing technology as well as alignment and variant calling algorithms, this isn’t that surprising.
What may be surprising is breaking these numbers down by Single Nucleotide Variants (SNVs), which we commonly think of as easy variants to call, and small Insertions/Deletions (InDels) which are known to be inherently more problematic.

Concordance of SNVs between the three service providers. Note both Illumina based sources have a ~80% concordance rate and Complete Genomics is calling far fewer variants in total.
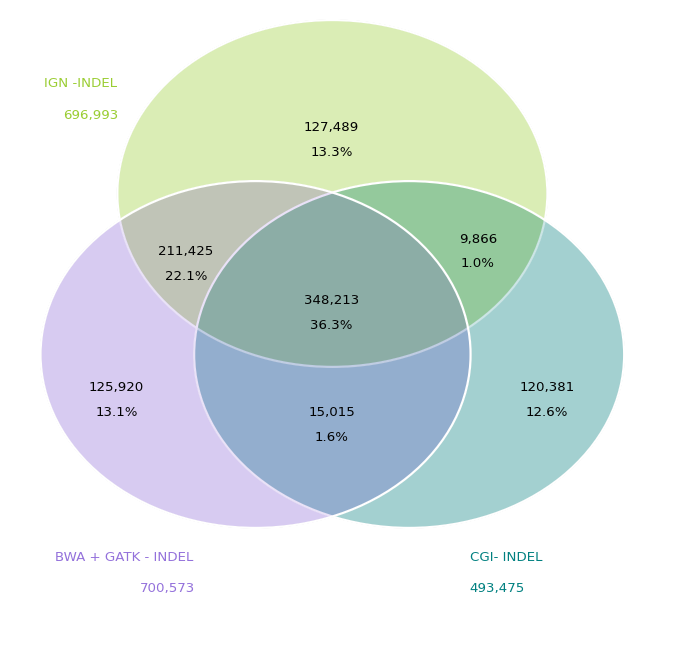
Concordance of InDels between groups based on matching start position and mutation type only. Here each source has roughly the same number of InDels that are uniquely called by themselves. Between the Illumina based sources the concordance is roughly 60%.
To be fair, there are a lot of variants in these sets that would be filtered out as too low depth or poor quality in most analysis scenarios. Many of the variants that are called by only a single source probably fall into that camp.
It’s a bit tricky though to pick filtering thresholds in a consistently fair way between these different sequencing depths and platforms. More importantly, for my purposes, or any benchmarking use case, how does one know which of these three opinions of a variant’s definition is the truth?
My favorite way to find false positives with knowable truths is to look at de Novo mutations.
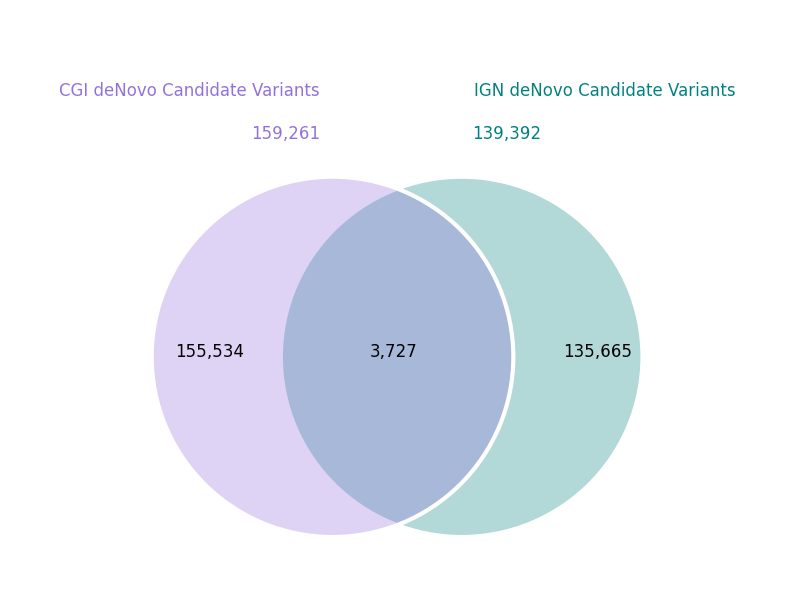
Overlap of de Novo heterozygous variants in NA12878. Although the overlap is small, 3,727 is still much much more than the expected number of true events.
I didn’t have the full trio for the BWA/GATK pipeline, but you can immediately get a feel for how much “noise” remains in these genomes by looking at the overlap of de Novo candidates between the IGN and Complete Genomics pipeline. Note, I’m narrowly defining de Novo events to just those sites where the child is heterozygous and both parents are homozygous reference.
Since it has been established that the number of true de Novo SNVs in a genome is less than 100, we can expect one or more of the three individual genotypes at each of those ~150k sites in each source to be called incorrectly (i.e. they are false-positives).
Even the ~4000 sites where the two sources agree, the number of events is highly inflated. Although one could argue we can expect a cell line like NA12878 to have a higher mutation rate than fresh germline samples, we are still an order of magnitude off here.
If we were to start to iterate through these ~4000 de Novo sites, we would see many of the enumerated confounders I listed above well exemplified.
Although I will go into this in more detail in Wednesday’s webinar, here is a great example:
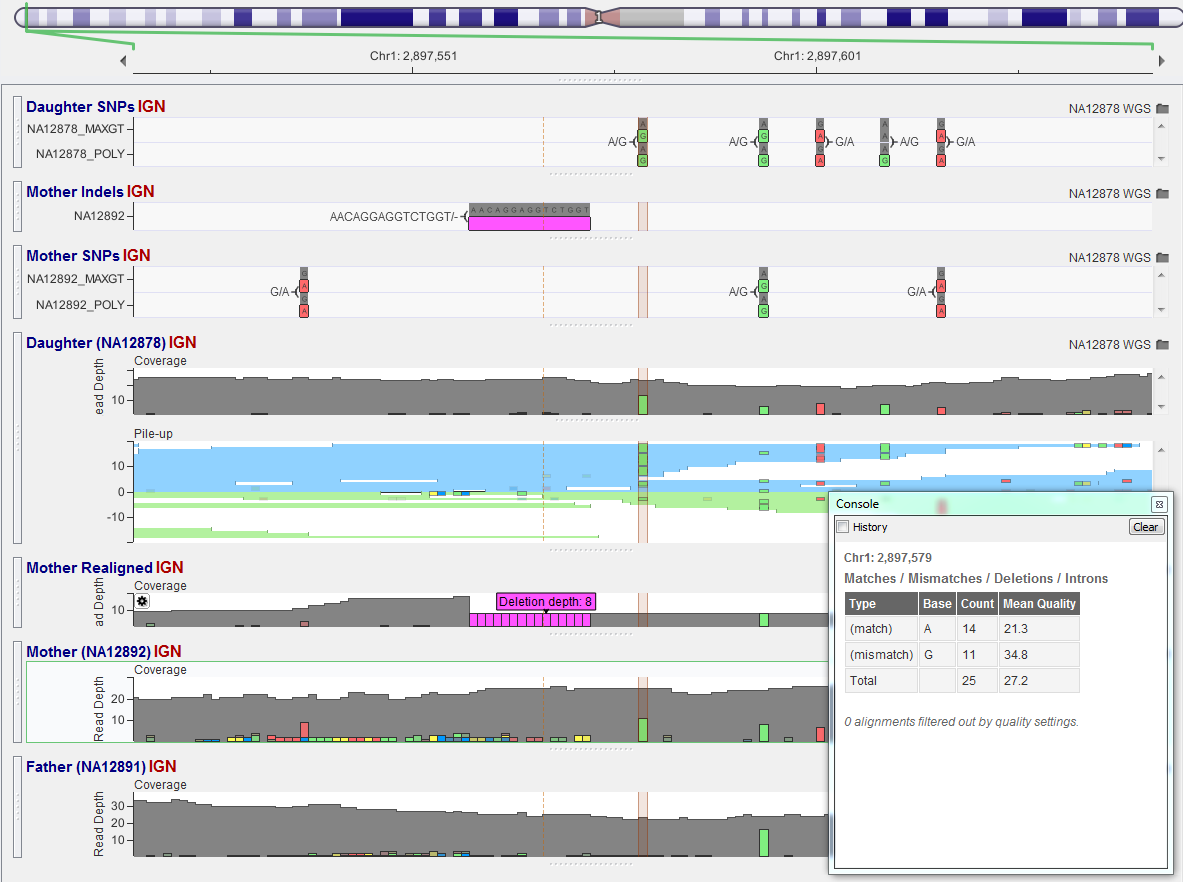
The daughter is called de Novo heterozygous, but clearly the mother is as well. It turns out the mother has a 8bp heterozygous deletion just upstream of this locus on the other chromosome. The variant caller was shy to call a variation so close to an InDel, but the data clearly shows it’s there.
But of course, you will also find some real de Novo mutations as well, such as this one.
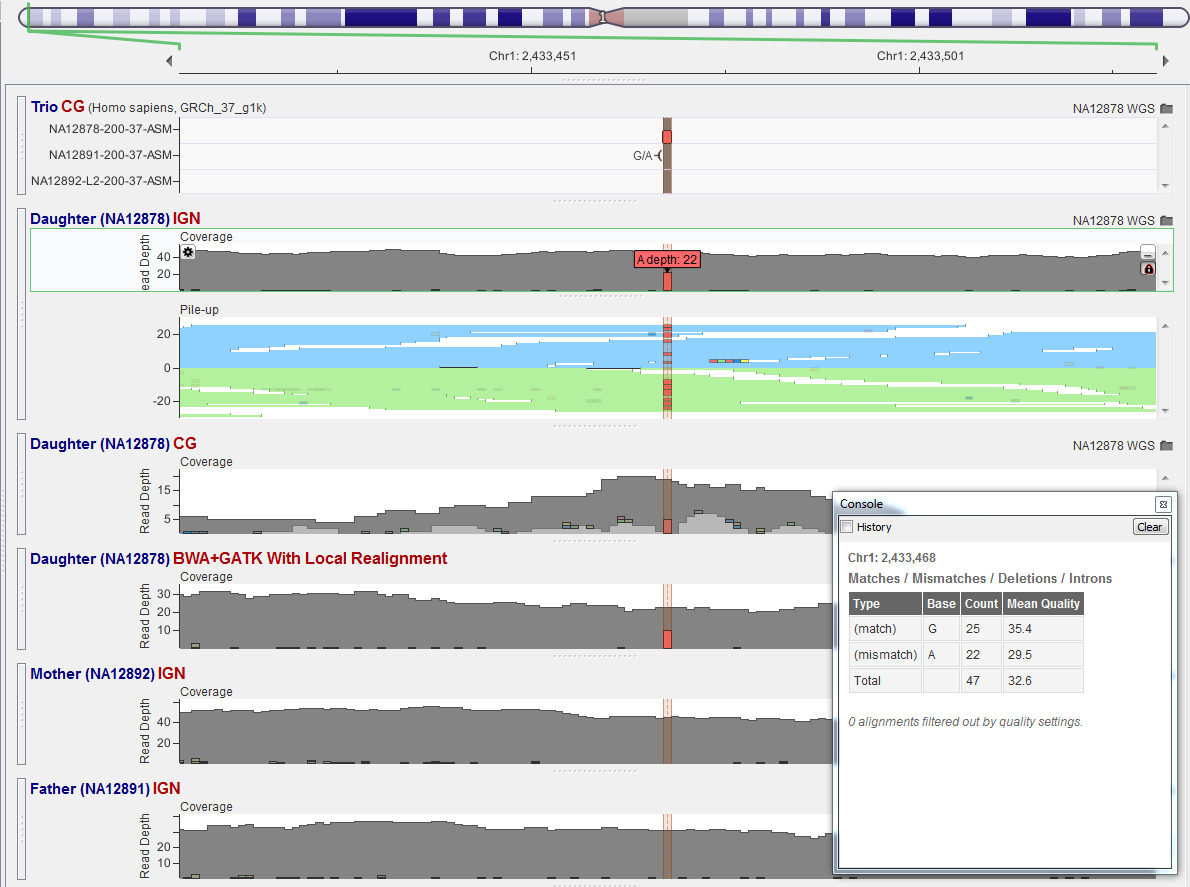
All three platforms called this as a heterozygous in the child. The Mother and Father don’t have a single read supporting the mismatch of an A. Squeaky clean de Novo mutation!
Don’t Panic, Just Bring Your Towel

By definition, we are looking at the worst and hardest to get right variants in these examples. It’s easy to feel a sense that these are lost causes. Hopeless cases.
But if I could inscribe some words on the cover of your newly minted genomes, they would be: DON’T PANIC.
While galactic hitchhikers may find a towel to be an indispensable utility, genome explorers should always be toting a genome browser as they come out of hyperspace and into a genomic coordinate of interest.
So the good news is, you can learn to spot these suckers a kilobase away with a good browser.
And the second piece of good news is that after years of frustratingly slow progress, these problem variants are being tackled by a whole new generation of offensive tactics.
Here are a couple that I’m aware of, maybe you can think of even more. If you can, please list them in the comments section.
- NIST is working on defining a “Genome in a Bottle” reference whole genome variant sets so we have both a gold standard variant set and common strategy in which to describe variations from the reference.
- The Genome Reference Consortium is coming out with GRCh38 this summer, applying roughly 200 patches to fix problematic regions of the genome.
- Additionally, GRCh is advocating thinking about the reference as a “graph” with multiple alternative paths for regions like the MHC with high population allelic diversity.
- The 1000 Genomes Project and others are thinking about classes of variants like multi-nucleotide variants as a vector of improvement.
- The GATK team is building their new variant calling framework in a haplotype aware model (called aptly the HaplotypeCaller).
- Read lengths are increasing and alignment algorithms like BWA-SW are being designed to handle them in ways that should improve detection of InDels.
- Long-read technologies such as Moleculo from Illumina could significantly improve calling of structural variants and other places with large scale divergences of a sample’s consensus sequence from the human reference.
- Start-ups like Real Time Genomics are focusing on the problem of “clinical grade” genomes and doing things like calling trio genotypes in a pedigree aware manner.
I don’t expect any of these individually to be a silver bullet. But in concert, I’m looking forward to some remarkable improvements in the coming year.
In the meantime, keep that genome browser handy and don’t panic.
Note: Thanks to Autumn Laughbaum for helping with the concordance analysis and graph generation.
Note 2: If you are just getting into genome exploration, a good start might be my Hitchhiker’s Guide to Next Generation Sequencing (Parts One, Two and Three). Yes, it may be equally laden with Douglas Adams references.
“Don’t panic” is a great thing to say also when delivering exome or whole-genome variant callsets to a collaborator who’s not familiar with NGS data.
As you point out, there are a number of groups and initiatives that should help address these “problem variants”. Personally, I think a new and more complete human genome reference has the potential to be one of the most valuable. Many of the false positives from variant calling are from the systematic mis-mapping of reads that came from a paralogous sequence not accurately represented in the current draft assembly. Build a better reference, and those reads will map properly. It stands to reason that false positive rates for indels and SVs would see a similar benefit.
While I think there will definitely be some variants cleaned up by GRCh38, there is only so much a better reference will give us.
Regardless of adding alternative loci for places like the MHC, there are plenty of regions with legitimate near-identical duplications in the human genome (see the “SuperDups” track in UCSC). With pseudogenes, mobile element insertions from retrotransposons and tandem repeats around, there are plenty sites that look messy when you align short reads to them.
But I am a believer in a better reference, but can you imagine how long it’s going to take for enough useful annotations to be lifted over or re-done on GRCh38 so that we can actually use it for analysis? We have orders of magnitude more data than we did for the NCBI36->GRCh37 transition! (Think ENCODE…)
Hi Gabe, thank you for all your summaries and explanations, they are very useful.
I was present at the San Diego conference on the morning of the 21 when NIST presented Genome in a bottle and talked about having control genomes. This is all good, but my impression is that it applies only to germline variants. However, as you well say, when you go into de-novo mutations the situation is much more complex. In fact, comparing cancer samples appears to me as a bigger challenge because of the presence of multiple clones. I have 2 questions:
1. What is the best way to compare 2 or multiple cancer samples or a cancer sample to the patient’s “normal” sample
2. I see that many tools accept only a vcf file. In this context, how confident can you be that a non-called variant is equivalent to the reference genome?
Thanks,
Hey Antonella,
Those are both excellent questions. I’ll hope to have time in my webinar to talk about calling somatic variants as you gain a lot by having the variant calling algorithm know it’s lookat for somatic variants in a tumor/normal pair. Depending on the clonal frequency of the tumor, you would want to call variants at much lower allelic frequencies of the alternate reads than you would for germline mutations.
I’ll also talk about the second question, but the answer is mixed. If you are looking at whole genomes, a site that is called confidently in one sample is almost certainly reference when not called in another sample as coverage is very consistent between samples. The alternative is that the sample without a variant has little or no read coverage and we can’t make a call. For exomes for example, this is more likely as coverage can vary more dramatically between samples (but again, for the most part a high quality call in one sample should be covered and reference in others).
The best solution is to have samples that need to be analyzed together be called together to create a multi-sample VCF file where each site that has a variant in one sample has the genotypes called (reference or no-call) in all samples.
Pingback: VarSeq: Making Variant Discovery and Gene Panels Easy | Our 2 SNPs…®
Pingback: Our top 5 most visited blog posts | Our 2 SNPs…®
Pingback: Comparing Variants using a Venn Diagram | Our 2 SNPs…®