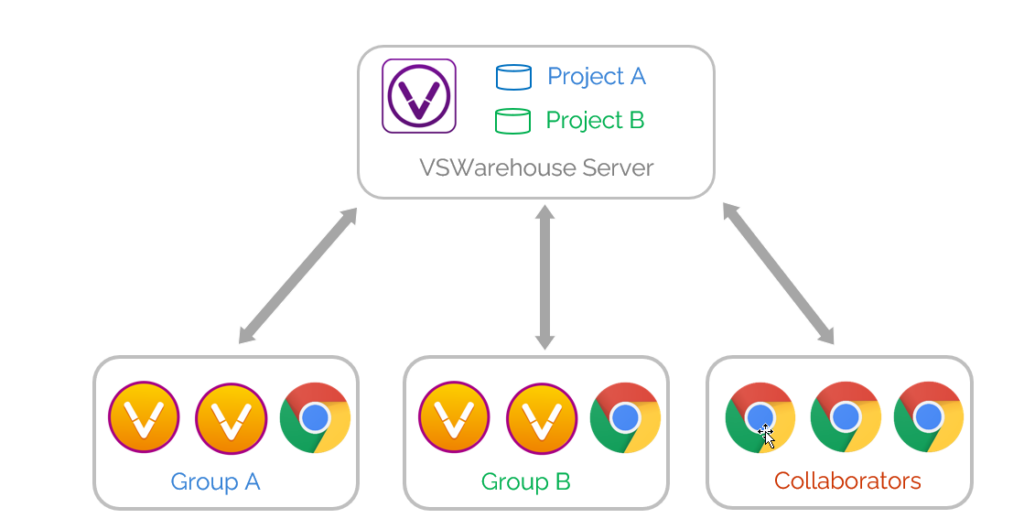
VarSeq Warehouse is the solution provided by Golden Helix for management of large-scale genomic data. It serves a centralized, indexed variant repository that stores variants and assessments from selected samples or projects. Management of large-scale genomic data with VSWarehouse allows entire teams, including collaborators, to manage various high level aspects of their NGS workflows, such as allele frequency tracking across cohorts, cataloging, reusing and sharing of variant assessments, genomic data querying, variant reanalysis, creating annotations, and integrating NGS data with EHR and LIMS platforms. This blog will review several use cases for VarSeq Warehouse.
Tracking Variant Allele Frequencies Across Cohorts
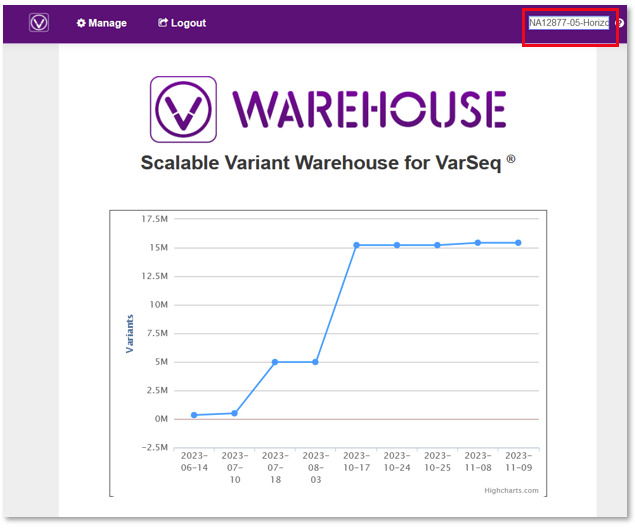
Organizations performing NGS analysis tend to scale up over time and inevitably accumulate large amounts of genomic data. This data is often used to gather useful insights on a cohort level but the management of the data as they scale up can pose some challenges. For example, users often need a structured framework to keep track of the allele frequency across batches of samples. This is important for tracking common variants in their specific study population, identifying sequencing artifacts and tracking variants that segregate with a particular disorder, among other use cases. VSWarehouse provides a solution for this by allowing users to upload entire projects spaced out in batches over time to a central Warehouse project, enabling the user to easily track the allele frequency of the growing cohort over time. To utilize VSWarehouse, users need to provide a physical server infrastructure to host their genomic data, enabling the user to have control over data safety, while Golden Helix provides the software framework uniquely designed for management of large-scale genomic data.
Querying Genomic Data
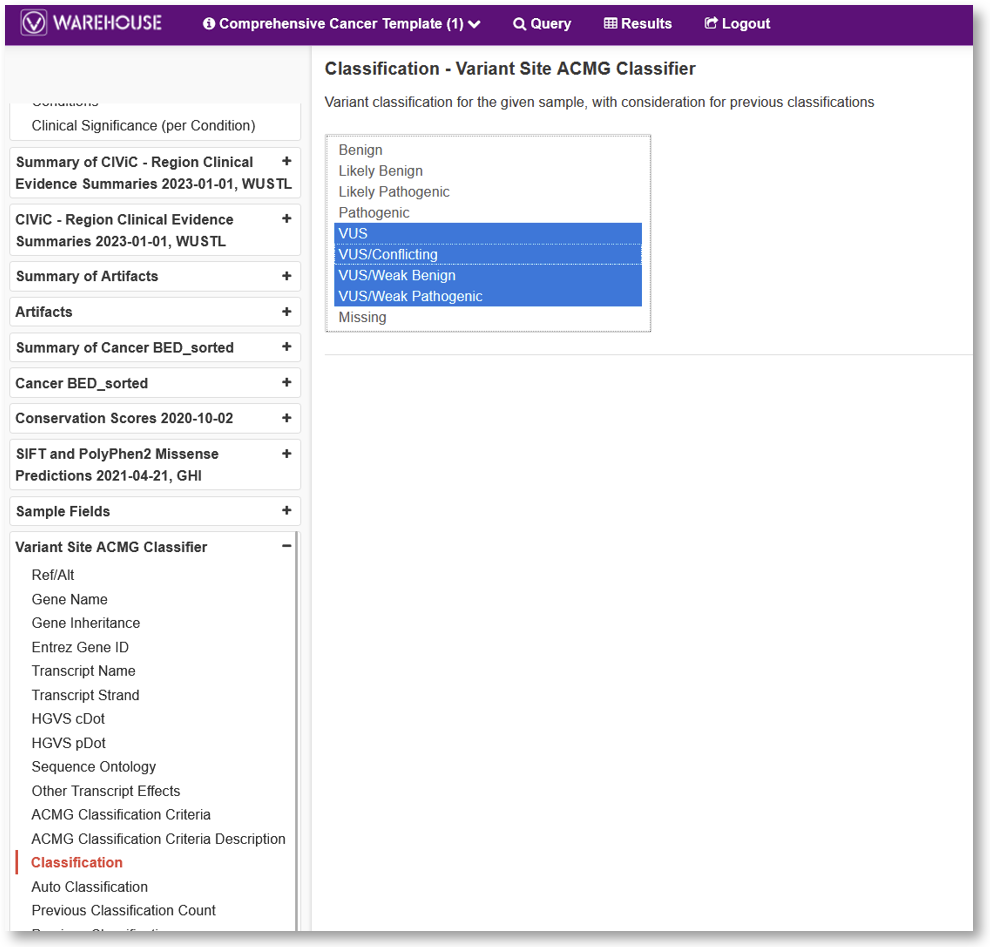
VSWarehouse places the user’s entire cohort of samples and variants at the user’s fingertips for querying. A VSWarehouse project will minimally include a gene track annotation such as RefSeq Genes but, of course, may include variants annotated with any other compatible annotation or algorithm that a user would want to be able to query in Warehouse. A very common use case is querying all the variants of uncertain significance (VUS) in a cohort of samples that were analyzed in a previous month or year for auditing and reclassification.
Variant Reanalysis and Reclassification
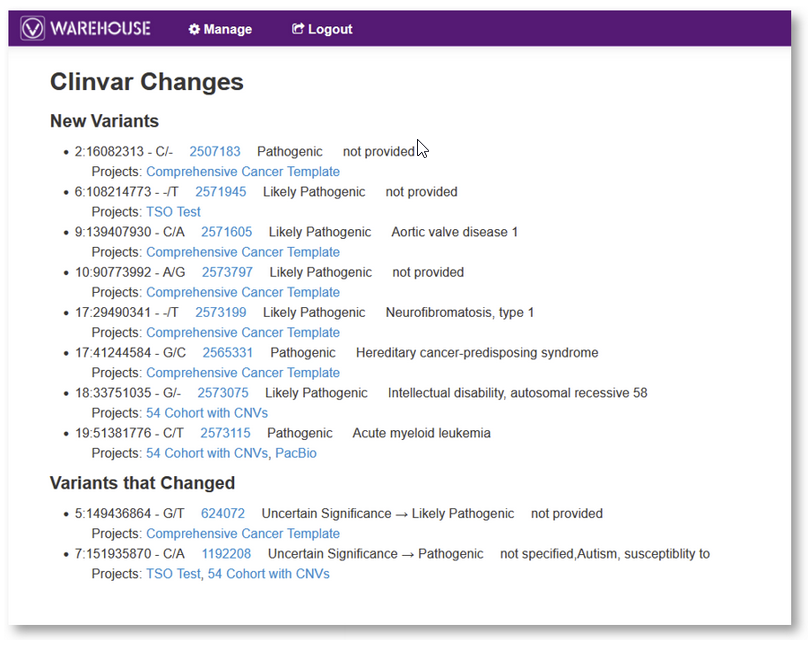
With regards to auditing and reclassification, VSWarehouse provides a very useful feature – the ClinVar Changes tracker – that allows users to quickly scan their entire collection of variants for updates in ClinVar classification. VSWarehouse will present the list of variants in the user’s projects that were previously unclassified and now have a ClinVar classification, as well as those that have a classification that has been updated to a different category.
Sharing and reusing variant assessments
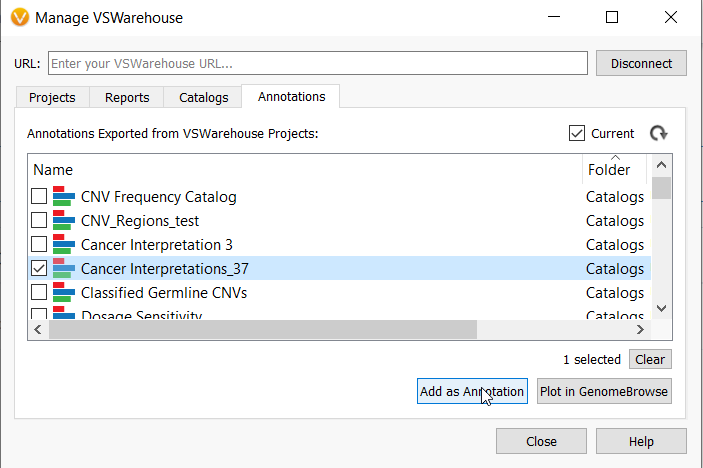
VarSeq Warehouse takes variant interpretation management for large genetic cohorts to another level. With Warehouse, users can access variant assessments and genomic data from any user on their team as long as they have been granted access. This genomic data can be used in a current project, for example, to plot and annotate with assessment catalogs stored in Warehouse by anyone on your team. This feature saves the entire team time, minimizes rework and boosts collaboration as you can reuse a variant assessment performed by other members of your team, or reuse your own variant assessments stored in your Warehouse catalogs over time.
Collaboration and Management of User Access
VSWarehouse is an enterprise level project management tool for large-scale genomic data which enables teams to manage different users’ access to information. An administrator of an organization’s Warehouse can define which users have access to which projects and who has the power to view as well as make changes to items stored in Warehouse such as projects and reports. These access management capabilities greatly facilitate collaboration within and across organizations.
Integrating Genomic Data with Patient Management System
VSWarehouse has a robust API that can be used to transfer data between VSWarehouse and EHR/LIMS systems. VSWarehouse is the web-based VarSeq resource which facilitates the transfer of genomic and sample data back and forth between VarSeq and EHR/LIMS systems, further enabling enterprise level management of large-scale genomic data.
Conclusion
VSWarehouse has numerous features that are useful for management of large-scale genomic data. This blog gave an overview of how VSWarehouse can be used in an NGS project management workflow. For more information on accessing VSWarehouse or if you have any comments and questions on the content of this article, please reach out to [email protected].