In our previous webcast, Evaluating CNVs with VSClinical’s New ACMG Guidelines, we focused on a CNV deletion (12:27715515-29628122×1) in which the patient had a known disorder called Brachydactyly type E. The CNV was isolated using our VS-CNV caller and applied to the ACMG CNV guidelines using the intuitive steps of VSClinical. If you missed the webcast, you can watch the on-demand video below.
One of the first steps in evaluating CNVs according to the ACMG guidelines is to determine if there are genes that the CNV overlaps that have dosage sensitivity information. As displayed in Figure 1, the algorithm identified that the PTHLH gene had some evidence of haploinsufficiency but not enough to be applied to Section 2 of the ACMG guidelines. Nonetheless, this gene was selected to be the critical gene to evaluate based on the number of available annotations including OMIM, Orphanet, and DHI.
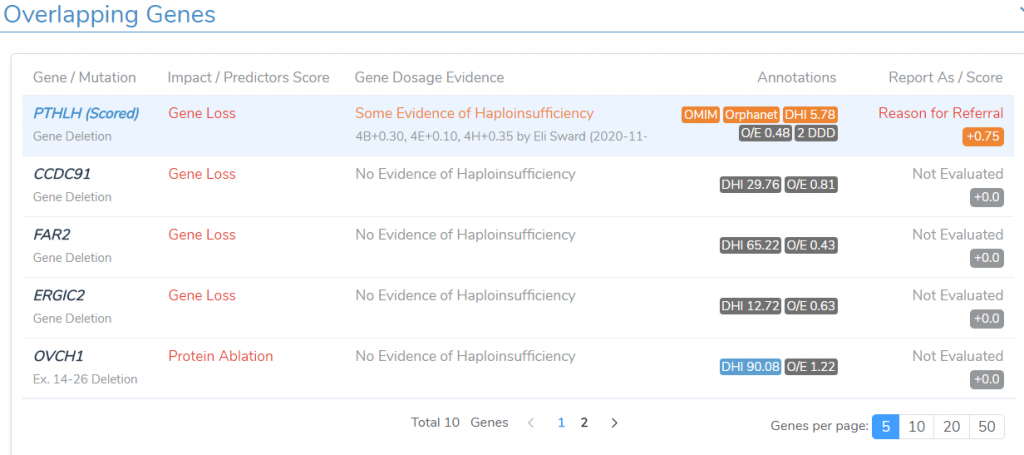
Figure 1: ACMG algorithm will identify the critical gene that the CNV event overlaps.
Because there was little evidence of dosage sensitivity, a literature search was required for relevant publications that could be applied to Section 4 of the ACMG guidelines in VSClinical. This section incorporates a detailed evaluation of genomic content using cases from published literature, public databases, and/or internal databases. Ultimately, the publications listed below are reports associating loss of function variants or CNV losses in PTHLH with Brachydactyly type E. Furthermore, valuable information regarding the inheritance of the mutation was documented for each publication.
Individual case evidence- de novo occurrences
- PMID: 26640227, c.101+3delAAGT (Gene Loss)
- PMID: 28211986, c.166C>T (p.R56*) (Gene Loss)
- PMID: 26763883, c.166C>T (p.R56*) (Gene Loss)
Individual case evidence- unknown inheritance
- PMID: 20170896, (p.K120X) (Gene Loss)
Individual case evidence- segregation among similarly affected family members
- PMID: 30458061, Frameshift (Gene Loss)
- PMID: 29947179, c.169C>T (p.Arg57*) (Gene Loss)
- PMID: 26640227, c.47_101þ73del128 (Gene Loss)
- PMID: 26763883, p.N87Tfs*18 (Gene Loss)
The reports were then applied to Section 4 of VSClinical using the intuitive decision trees, which was demonstrated in the webcast. The result was the summation of the publications, which was 0.75, as shown in the image below. As you can see these resulted in the PTHLH gene containing some evidence for haploinsufficiency, which could then be used in the total point scoring system for this event.
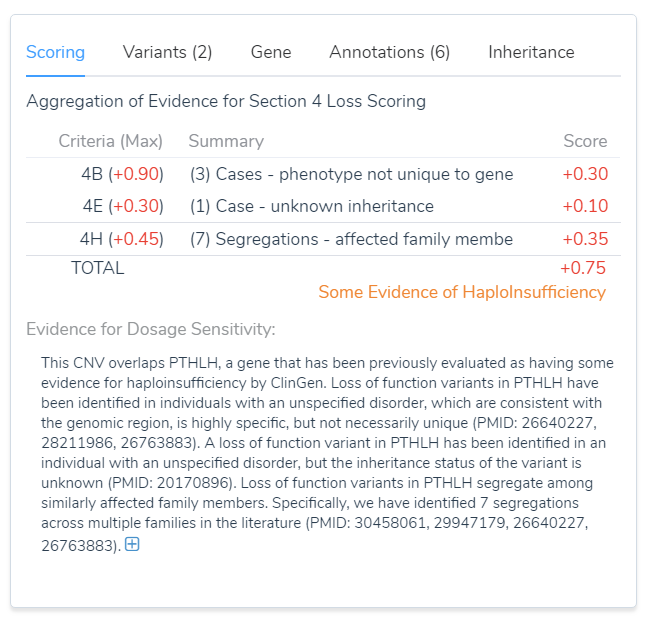
Figure 2: Using Section 4 we were able to reach a score of 0.75 and some evidence of dosage sensitivity.
In this webinar, we also covered Section 5 which is based on the evaluation of inheritance pattern/family history for the patient being studied. For this example, we assumed it was a de novo inheritance pattern and that the phenotype was highly specific, but not necessarily unique. This produces an additional 0.15 points to reach a final score of 0.9 and a Likely Pathogenic classification, Figure 3.
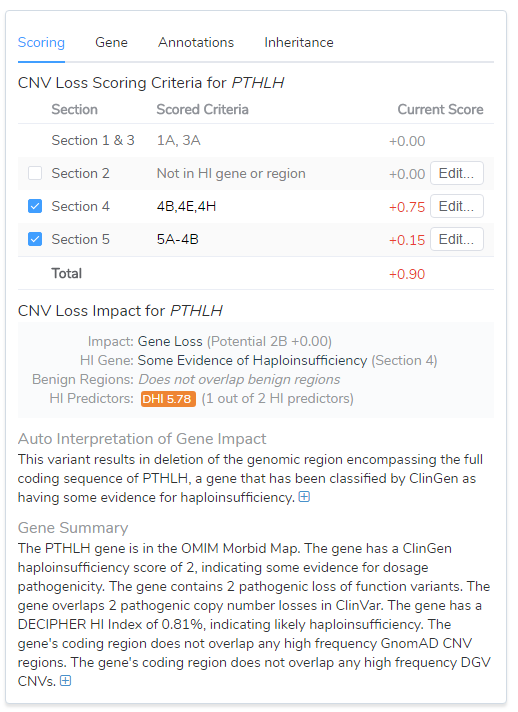
Figure 3: Combining Sections 4 and 5 resulted in a Likely Pathogenic classification.
We then ended the demonstration with a clinical report and showed the automation of this process. The interpretations were generated automatically from the criteria that was answered and was saved in internal assessment catalogs for future evaluations. In this demonstration, we did not have time to fully cover the reporting capabilities but if you are interested in the report generation and customization, you can register for our next webinar. This upcoming webinar will also go into other details of the VSClinical ACMG CNV interface and show how easy it is to make customizable reports.
I hope you enjoyed reading a brief synopsis of the webinar and if you would like to see more you can watch the full recording here. Feel free to check out some of our other blogs that always contain important news and updates for the next-gen sequencing community. As always, if you have any questions about this webinar or would like to have a demonstration or evaluation of the software please contact us at [email protected].
