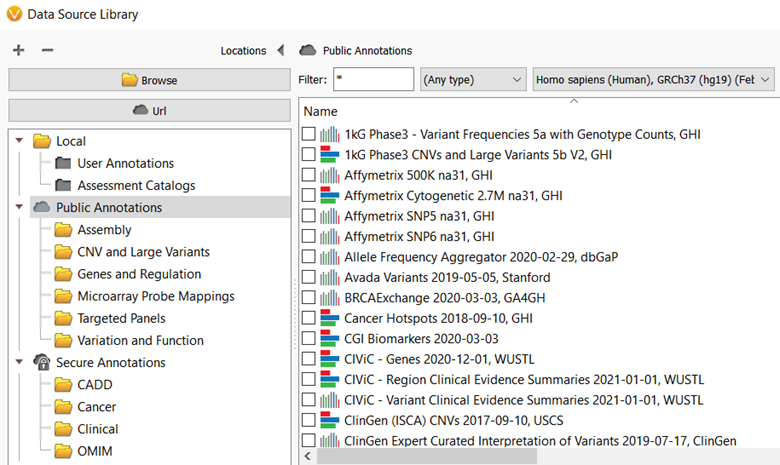
Annotating genomic variants is a very complex process but perhaps the most important part of next-generation sequencing variant analysis. Here at Golden Helix, we recognize the importance and value of having the most up-to-date sources available and curating new annotation sources as they become available for variant analysis. Golden Helix has curated over 100 annotation sources for human variant analysis and even more, various annotation sources are used by algorithms in the software for an added application. Many of these annotations are available to all VarSeq and SVS users through the Public Annotations server (figure 1). However, in this blog, I want to talk about another annotation server that you may not know about, the Staging Annotations server!
What is the Staging Annotation server used for? In general, there are two reasons to use annotations hosted on this server:
- To access a user requested annotation source
- To access an annotation that is not publicly available and released in the software yet
Let’s start with reason number one. As many of you already know, we are always open to feature requests to improve our software. In some cases, customers have requested databases that they would like curated and incorporated into the software as annotations. As straightforward as this may sound, there are some cases wherein curating the database for use in VarSeq or SVS has some issues or caveats for use and does not apply to each user’s workflow. Often people that are familiar with the database are aware of these issues or caveats and still would like to use the source in VarSeq or SVS.
Reason number two occurs when the annotation has been curated, but it is not fully integrated into the software yet. This is the case when there is an algorithm that uses the annotation source for computation. A great example of an annotation that is used in many of the algorithms is RefSeq Genes, as often the gene names are used by the algorithm. However, we often make these sources available for users to add to their projects until we fully integrate the annotation within the software by storing them on the Staging Annotations server.
There are certainly some important considerations when using annotations on the Staging Annotation server which I will discuss momentarily, but first I want to show how users can access the server. The Staging Annotations server can be added to the Data Source Library by clicking on the plus icon in the top left-hand corner of the Data Source Manager window (figure 2). To open the Data Source Manger, go to Tools–>Manage Data Sources.
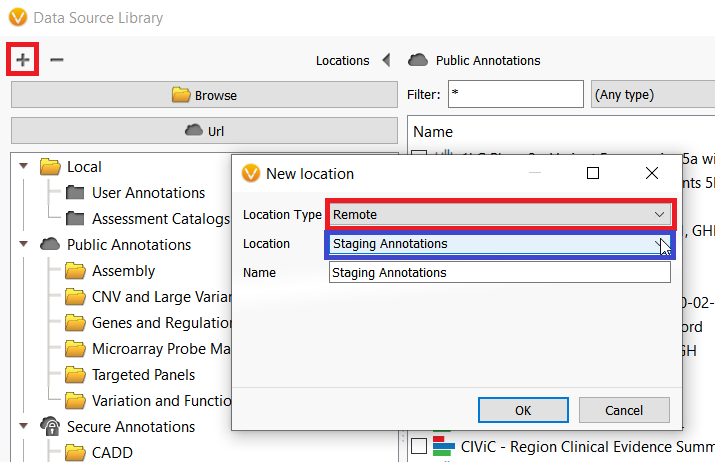
In the next dialogue, select Remote from the Location Type dropdown menu and Staging Annotations from the Location dropdown menu and click OK (figure 2). After adding the server, these annotations can now be downloaded and used in your project.
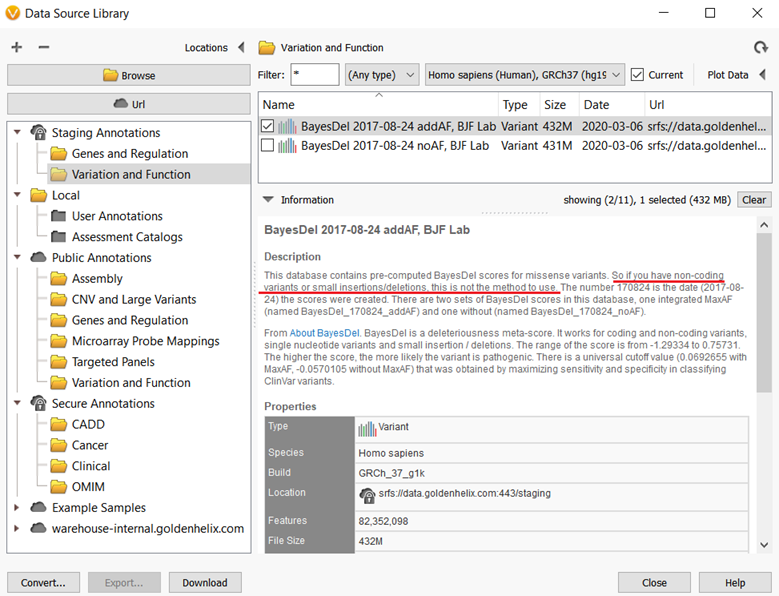
Figure 3 shows an example of an annotation that was requested by a Golden Helix customer, the BayesDel predictions for missense variants. Anyone can use this source for variant analysis thanks to the Staging Annotation server, however, there are reasons users should be cautious when using some annotations from the Staging server. If you are not familiar with the BayesDel predictions, you might miss a caveat with applying the source to your project. In this case, the BayesDel predictions score only for missense variants and this method should be used for small insertions/deletions or non-coding variants.
The second consideration users should be aware of when using an annotation from the Staging server is regarding the scenario that the annotation has not been fully incorporated into the software yet. I mentioned that the annotations may be on the server because they will impact algorithms used in the software, I used RefSeq as an example earlier. In some cases, adjustments are still being made to the annotations to fit the requirements of the algorithm, so they are still somewhat “a work in progress”.
That being said, there are advantages to using annotations from the Staging Annotations server in that you may be able gain access to an annotation source that you requested to be curated or you can gain access to the latest version of an annotation source and not have to wait for a new version of the software to be released.
If you have questions regarding the annotations on the Staging Annotations server or if you have an annotation source that you want to use and submit a feature request, please reach out to us at [email protected] and we are happy to help! Feel free to also check out some of our other blogs that always contain important news and updates for the next-gen sequencing community.