Detecting CNVs from whole genome data has a number of advantages but also unique challenges. Whole genomes offer a comprehensive and uniform picture of the entire genome, allowing a user to capture CNVs at a higher resolution than with data sequenced at a lesser scale. It also allows for the detection of structural variation over non-coding regions and for a more accurate definition of breakpoints.
One downside with whole genome CNV calling is it can become expensive, but using the VarSeq CNV caller allows precise detection of CNVs on shallow whole genomes, thereby reducing the potential cost of implementing this assay into NGS workflows. This blog explores how VarSeq CNV caller uses bin size adjustment to manage sensitivity and manage CNV calling strategies for detecting events of the desired scale.
Managing Sensitivity
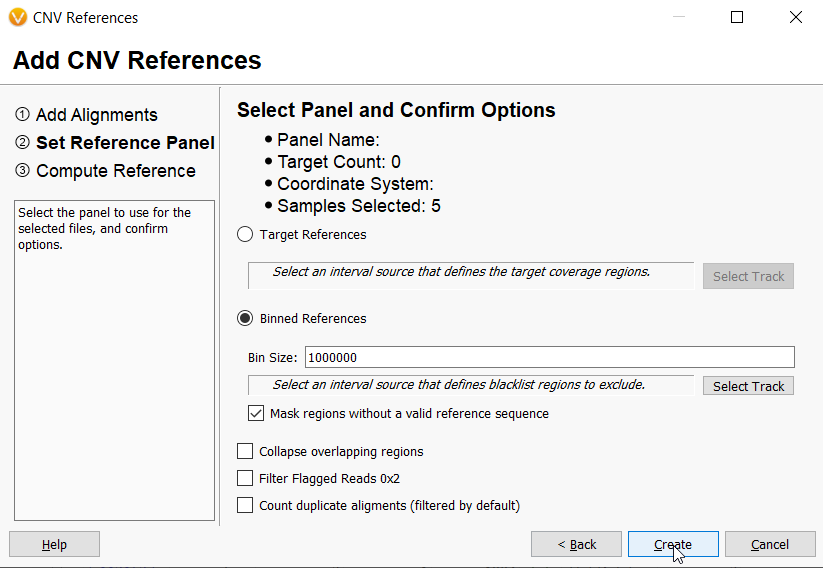
The Binned CNV Caller is designed for detecting large CNV events usually in a whole genome sample setting. Contrast this with our Targeted CNV caller, which is more suitable for calling CNVs down to the level of an exon in small gene panels and exomes, and requires a BED file to define specific targets on which to call CNVs. The default bin size for the Binned CNV caller algorithm is 1,000,000 bases and is ideal for the detection of large cytogenetic events in shallow whole genomes. However, we have made our Binned CNV caller settings loose enough to accommodate other use cases. In fact, users can set bin sizes ranging from 10,000 to 1,000,000,000. Smaller bin sizes will allow you to detect smaller events, thereby increasing the sensitivity of the caller. However, the binned caller will not detect CNVs that are smaller than several bins in size. Since the smallest bin possible is 10,000, the binned caller is limited to calling CNVs that are at least tens of thousands of bases in size. Someone doing, for example, hereditary breast cancer CNV analysis, who is expecting to detect CNVs within the BRCA1 gene, for example, would not want to use a bin size of 1,000,000 since the BRCA1 gene is only 110kb in size. For such an application a bin size of 10,000 may be more applicable. However, when choosing bin sizes, it is important to keep in mind the tradeoff between increased sensitivity, longer run times, and higher coverage requirements.
Managing References
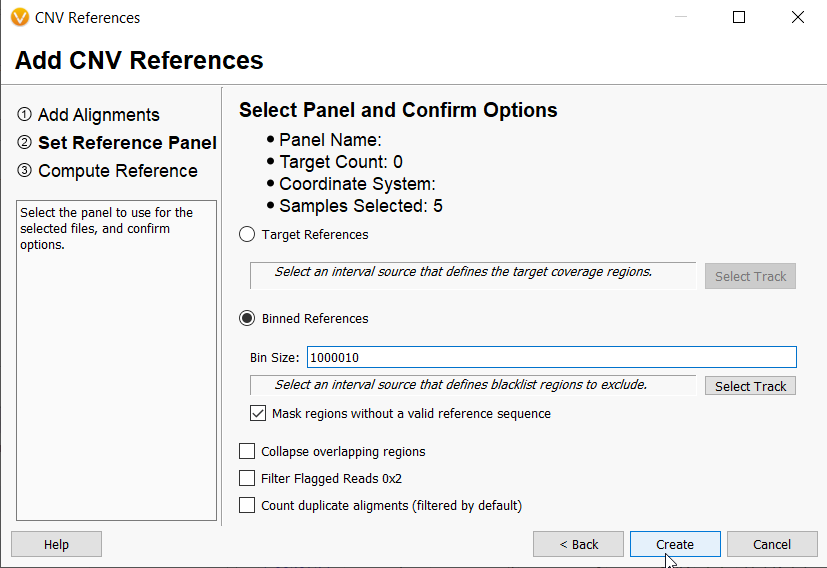
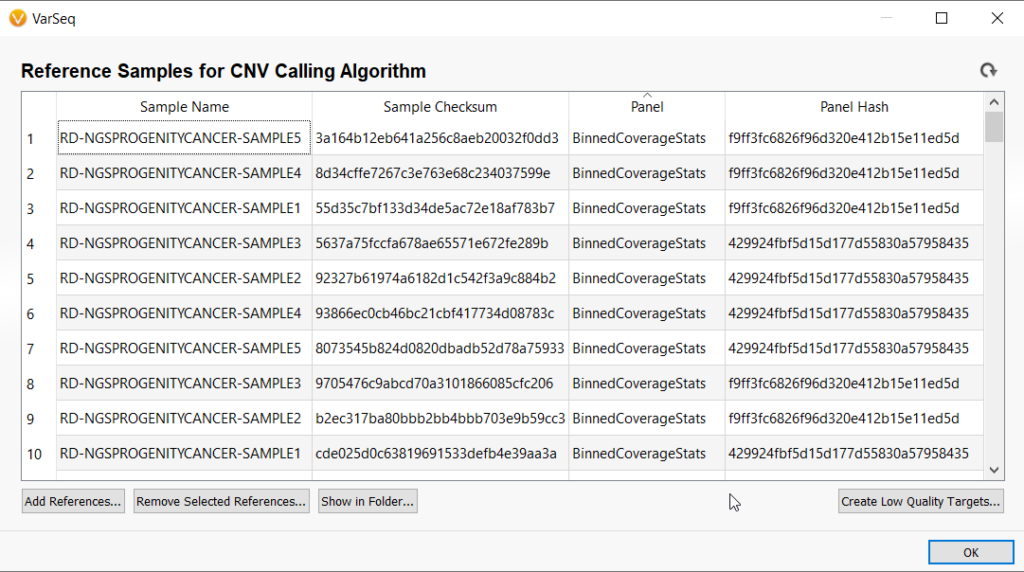
The VarSeq CNV caller uses reference sets as the basis for CNV calling on samples of interest. When managing references for multiple use cases, it is easy to define specific use cases for the Targeted CNV caller by using different BED files. Reference set management when using the Binned CNV Caller is also quite simple. To define a unique reference set for a unique use case, a user simply needs to make an adjustment, however large or small, to the bin size used for calculating coverage. In our example below, you can imagine that a user has a CNV reference set already run at 1,000,000 base pairs. However, they have received another batch of samples that were run with a vastly different sequencing method for example, and they need to create a separate reference set but also want to use bin sizes in the 1,000,000 range.
They can make a slight adjustment (setting the bin size to 1,000,010 in this case) to create a separate reference set.
I hope this article has shed light on what to consider when choosing bin sizes for the VarSeq Binned CNV caller. Please email us at [email protected] with any questions or comments on this topic. Thank you!