Clinical Variant Analysis for Cancer – Applying AMP Guidelines to Analyze Somatic Variants
Detecting cancer at an early stage can make it much more treatable. Developing tests and making them clinically actionable is crucial to beat this disease. This eBook covers the state-of-the-art gene panel tests for cancer. Of course, there is more that can be done. The field is pushing into exome, whole genome and RNA sequencing to capture more of the complex processes behind oncogenesis.
With VarSeq, Golden Helix has developed a software platform that supports gene panel analysis leading to clinically actionable information. This covers all the intermediate steps to reach a set of high-quality reportable variants, including:
- Setting the patient’s tumor type and other laboratory information
- Review the NGS panel statistics and quality metrics for the current sample
- Checking and reporting target regions that fail the thresholds set by the lab test
- Confirm that important must-call hotspot regions and variants have sufficient coverage
- Adding somatic and potential germline variants to the analysis, including SNV, CNV, and Fusions
- Scoring variants that are novel or of uncertain impact to a cancer gene before reporting it as a Biomarker
- Writing interpretations following the AMP Tier guidelines that incorporate the supporting clinical evidence for a biomarker in the context of the current patient’s tumor type
- Reviewing and finalizing the clinical report
In this example, we will review the results of an NGS gene panel covering
common cancer genes with a patient diagnosis of Melanoma. We will follow the
process from start to finish for the BRAF V600E biomarker and then cover a CNV
and Fusion example.
Patient Clinical Information and Tumor Type
At the start of the workflow, the Patient screen provides inputs for various patient information that may need to be displayed on the final clinical report.
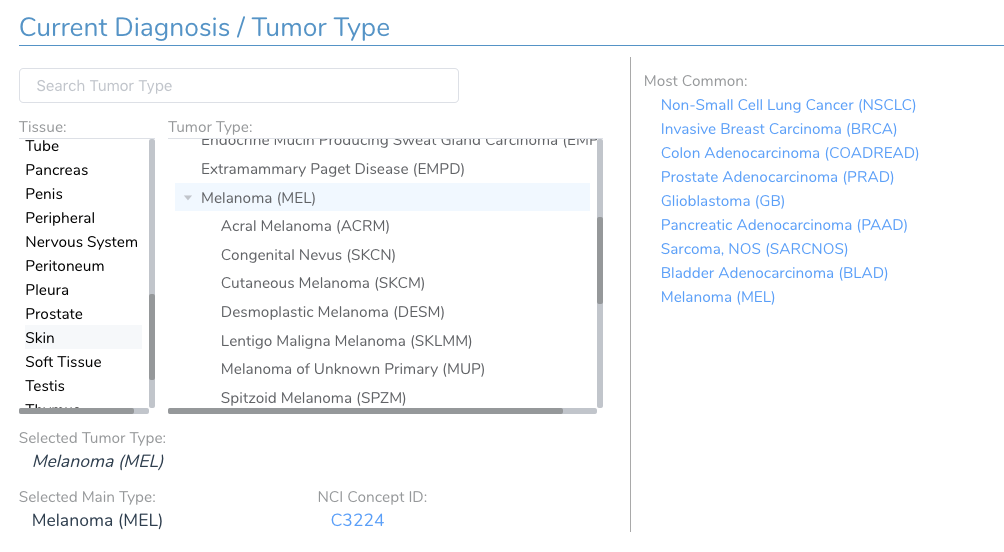
One critical patient specific field that is set in this first screen is the current diagnosis of the patient. The AMP guidelines emphasize the evaluation of clinical evidence for a cancer biomarker should be made in the context of the patient’s tumor type. The most intuitive way to represent the many cancer types and sub-types is through a tumor type hierarchy organized first by the originating tissue type. A searchable tree-view of the cancer ontology is provided to select the appropriate tumor type. Commonly tested tumor types are available for quick selection. Figure 13 shows the selection of Melanoma for this patient.
NGS Panel Summary and Quality Check
Also, in this first screen is the NGS Sequencing Summary and NGS Coverage of Genes in Figure 14. The critical decision can be made with these summary statistics on whether the sample needs to be re-processed or whether it meets the standards of the test to be used for clinical assessment.
If you wish to continue reading the eBook, I invite you to download a complimentary copy of my eBook. You can do so by clicking on the button below.