VSWarehouse 3 (VSW3) is a powerful genomic data warehousing solution that gives laboratories full control over their cloud deployments while ensuring security and flexibility. One of its most exciting new features is the ability to run custom applications directly from the browser. This means that users can launch specialized tools to analyze and visualize genomic data without leaving their secure cloud environment.
VSWarehouse 3 uses this capability to integrate VarSeq as an application that can run directly in the browser, with direct access to the projects and genomic data hosted on the server. But it is also possible to add bundle and run any Linux or web application on the power application platform!
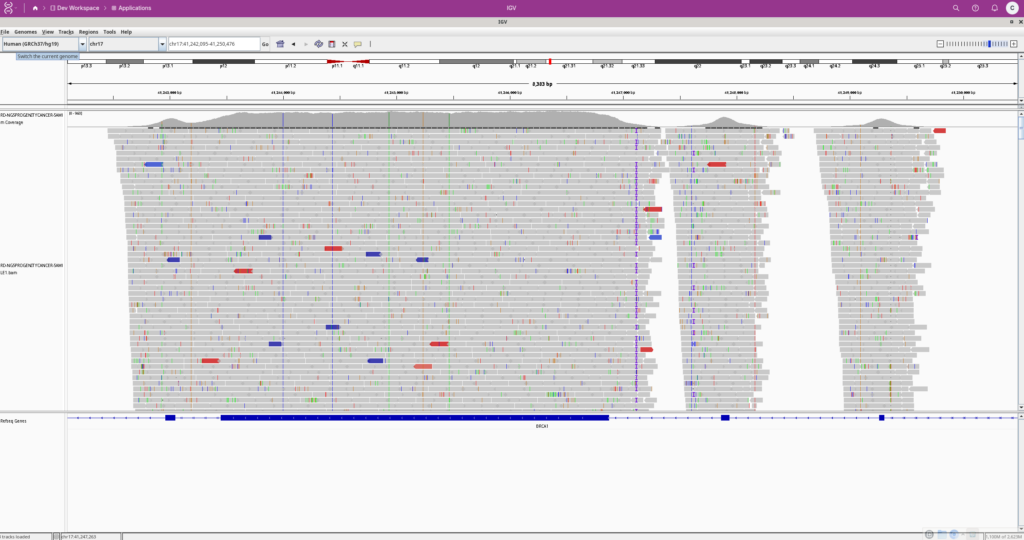
In this post, we will explore how users can leverage the VSWarehouse 3 application functionality to run the Integrative Genomics Viewer (IGV) directly on the server. IGV is a widely used genome browser that allows users to visualize BAM, BED, and VCF files, providing an interactive way to explore genomic data.
How Custom Applications Work in VSWarehouse 3
VSW3 allows users to define and run any application that can operate on Linux or through an HTTPS server. Applications are defined using a YAML configuration file, which specifies details such as the required computing resources, input parameters, and the Docker image that will be used to run the application.
Each application in VSW3 is run inside a Docker container, ensuring consistency and isolation from other applications. These Docker images can be stored in a public repository like Docker Hub or a private repository for added security. For ease of use, Golden Helix provides a base image that can be modified to run any Linux application. This means users can quickly set up their own tools without extensive configuration.
Deploying a custom application on VSW3 involves two steps:
- Build a custom Docker image: A Docker image is a lightweight, standalone package that includes everything needed to run a piece of software, ensuring it works consistently across different environments. Each VSW3 application has an associated Docker image that defines the application enviroment and is accompanied by a script specifying how the application will be launched.
- Define the application using a YAML file: Each application on the warehouse has an associated yaml configuration file that specifies the application details, including the parameters and resource requirements.
While diving into the details of how to define and build Docker images is beyond the scope of this blog, I do want to spend some time going over the YAML configuration files in the context of our IGV application.
Setting Up IGV as a Custom Application
The YAML configuration file below defines the parameters for our custom IGV application:
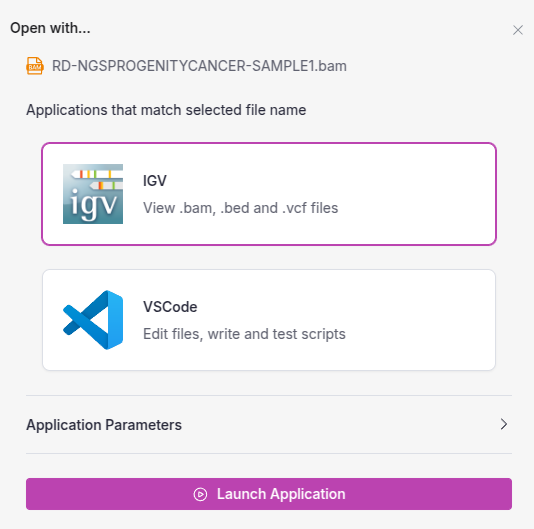
name: IGV
description: View .bam, .bed and .vcf files
icon_path: images/igv.png
agent_requirements:
cpu_cores: 2
memory_gb: 4
parameters:
- name: IGV_FILE
label: IGV File
type: file
optional: true
pattern_match:
- '*.bam'
- '*.bed'
- '*.vcf'
- name: IDLE_TIMEOUT
label: Idle Auto Shutdown
help: If the user does not interact with the application for this amount of time, it will shut down.
type: enum
value: "28800"
choices:
"<1800>: Shutdown after idle for 30 minutes"
"<3600>: Shutdown after idle for 1 hour"
"<7200>: Shutdown after idle for 2 hours"
"<14400>: Shutdown after idle for 4 hours"
"<28800>: Shutdown after idle for 8 hours"
"<86400>: Shutdown after idle for 24 hours"
"<864000>: Shutdown after idle for 10 days"
"<>: Do not shutdown"
application:
docker:
image: registry.goldenhelix.com/public/ghdesktop-igv:2.16.0
workspace_mount: /home/ghuser/Workspace/
shared_memory_size: 1g
env:
VNC_PW: ${APPLICATION_PASSWORD}
NO_VNC_PORT: ${APPLICATION_PORT}
APP_ARGS: '"${IGV_FILE}"'
MAXIMIZE: 'true'
MONITOR_PROCESS: '1'The first block of this file defines basic information about our application, including the name, description, and icon. The second block specifies the application’s resource requirements, including CPU and memory limits that are enforced by the Docker container.
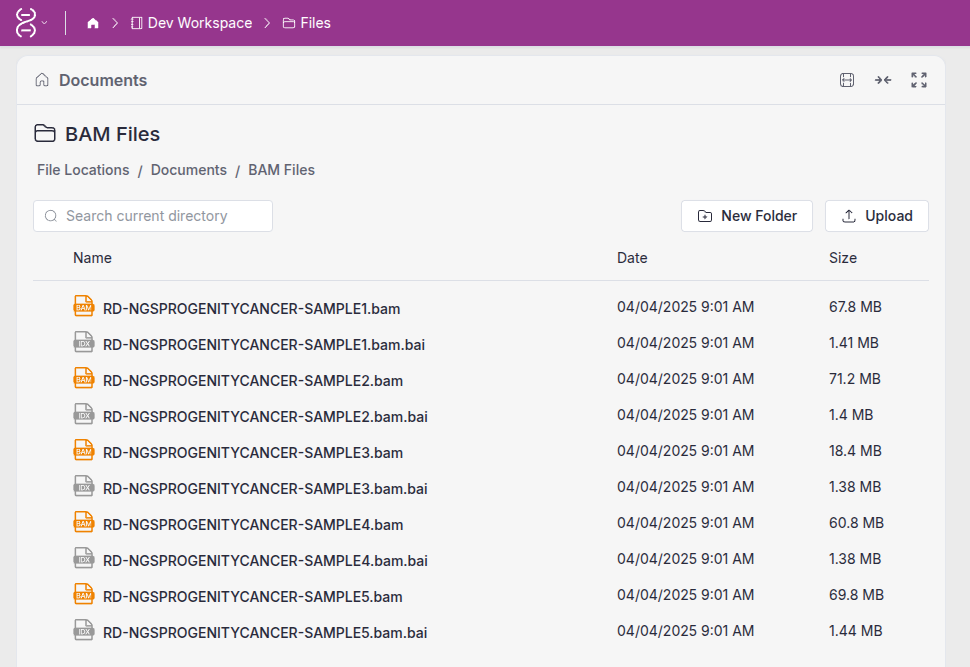
The third block defines application parameters. The IGV File parameter defines the type of files that can be opened by this application. This allows users to view files with these extensions in IGV directly from the VSW3 file browser.
Our IGV application now has complete access to all genomic data files within the workspace, allowing our users to seamlessly open and analyze multiple datasets at once.
The Idle Auto Shutdown parameter defines how long the application is permitted to be idle before being shutdown. The third block is the application schema, which defines the Docker configuration and environment settings. Each application must have an associated docker image used to launch the application, which is defined in the docker sub-section. The env sub-section is used to define any additional environment variables required by the application.
Once IGV has been added to VSW3, launching it is as simple as selecting the application from the interface and selecting the genomic data file for visualization. Users can interact with IGV just as they would on a local machine, but with the added benefits of cloud-based scalability and security.
Expanding Beyond IGV
While IGV is a great example, the custom application framework in VSW3 is designed to support a wide range of tools. Any Linux-compatible application can be containerized and launched within VSW3, providing laboratories with the flexibility to integrate their preferred analysis workflows directly into their cloud environment.
Additionally, while applications can be run locally or remotely, all communication with remote applications is performed through a secure encrypted and authenticated connection. This ensures that sensitive genomic data remains protected at all times, even when accessed remotely.
Administrators can create their own Docker images or modify the provided base image to suit their needs. Whether it’s a visualization tool, an annotation pipeline, or an AI application, VSW3 enables users to bring their custom tools to the cloud without compromising security or control.
Conclusion
With VSWarehouse 3, laboratories can seamlessly integrate their favorite applications into a secure and scalable cloud environment. The ability to define and run custom applications, like IGV, ensures that users have the flexibility they need while maintaining full control over their data. Contact us at [email protected] to learn more about VSW3 and how you can leverage its powerful new capabilities.