VarSeq enables users to import structural variants for annotation, filtration, and subsequent clinical or other analyses. Structural variants are often called during secondary analysis as belonging to two broad categories – Copy Number Variants (CNVs) with the file suffix “…_CNV.vcf” and Breakends with the file suffix “…_SV.vcf”. This blog will give some insight into how structural variants are triaged when imported into VarSeq, such that certain types of structural variants can be analyzed as breakends or fusions but also as copy number variants. I highlight a few ways in which users can take advantage of this superposition of specific structural variants for their analyses.
CNVs in Two Places at Once
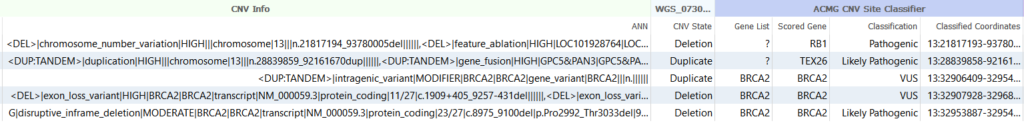
Is it a breakend? Is it a CNV? Yes. CNVs typically include deletions and duplications usually greater than 50 base pairs in length, while Breakends or Fusions include translocations, inversions and, yes, certain duplications, like tandem duplications, and deletions. As such, deletion and duplication events brought into VarSeq from structural variant (_SV.vcf) files may show up as both fusions and copy number variants. This mysterious phenomenon is no accident and can actually be used to your advantage. Below, I list two such ways:
Leverage the VarSeq CNV algorithms for Deletion and Duplication Events in SV files
The breakends that also qualify as CNVs have more algorithms that can be applied to them. At present, it is going to be much more informative to analyze, for example, tandem duplications in SV files as CNVs than as breakends. A major reason for that is because there are more annotations and databases currently in the field that describe CNVs than there are that describe breakends. Figure 2 shows the ACMG classification of tandem repeats in BRCA2, TEX26 and RB1, some of which are classified as pathogenic or likely pathogenic, which would have been unclassified as breakends. When using VarSeq, this triaging of such events is done automatically, so the user will not miss out on properly classifying these important events.
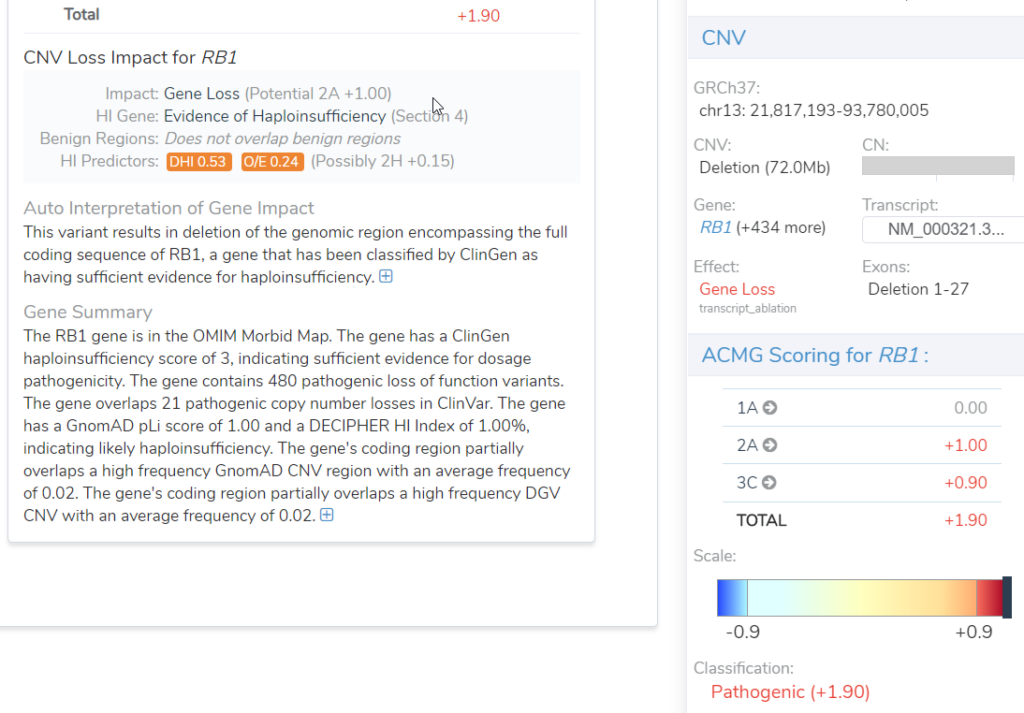
Leverage VSClinical CNV analysis for Deletion and Duplication Events in SV files
Importing such events as CNVs into VSClinical enables the user to leverage the ACMG ClinGen guidelines, while there are not yet any such guidelines for fusions. This allows the user to take advantage of the ClinGen records on haploinsufficiency and of course, our automated classification of the events in VSClinical (Figure 3).
Please let us know at [email protected] if you have questions about how to navigate these structural variant workflows in VarSeq. We would be happy to offer guidance on this and other related topics.