We have had many customers come to us over the years with a simple problem: they have BAM files for whole exome or gene panel data and would like to call CNVs using VarSeq’s powerful CNV calling capabilities, but they don’t have a bed file defining the target regions for their samples. To address this problem, we have developed a new feature that allows users to export any gene track as an exome region bed file, which can then be used to define the targets for the CNV caller.
To utilize this feature, select “Manage Data Sources” from VarSeq’s “Tools” menu and search for “RefSeq” in your local annotation tracks. Next, right-click the RefSeq gene track and select “Export.” You will then see this dialog:
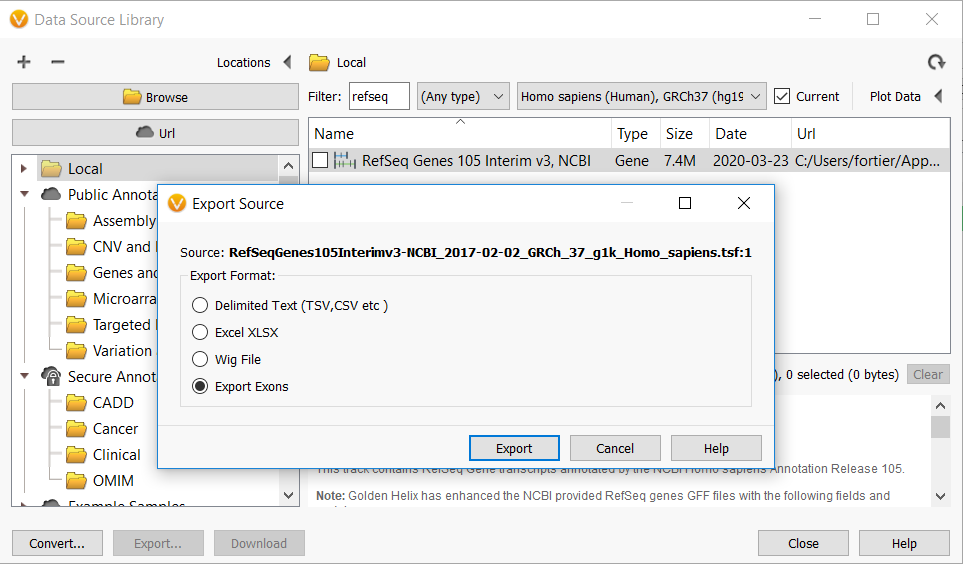
If you are familiar with our export functionality, you will notice the addition of a new option entitled “Export Exons.” After selecting this option and clicking “Export,” you will be given a series of options for exporting the exons of the gene track.

While the default options work well for most use cases, this functionality is highly customizable. Let’s explore some of these options to see how they can be used.
- The first set of options allows you to specify the padding around each exon and whether UTRs should be included in the exported regions.
- The next option allows you to choose which transcripts are exported. For CNV calling, it is generally recommended that you only export the clinically relevant transcript for each gene to ensure that there is only a single target region associated with each exon.
- Finally, you can choose to filter down to a sub-set of genes. This is useful if you are calling CNVs from gene panel data with coverage limited to a specific set of genes.
Once you are satisfied with the export options, you can click the “Browse” button to choose a location in which to save the file and click “Export” to save the newly created bed file. Now that you have a bed file specifying the target regions of your sample, you are ready to start calling CNVs.

If you want to learn more about VarSeq’s CNV calling capabilities, here are some of our other resources I recommend checking out!
- Great blog to provide an overview of the tool: VS-CNV; Golden Helix’s solution to replace traditional methods
- One of our many eBooks, free to download: Secondary Analysis – Calling Single Nucleotide and Copy Number Variants in Next-Gen Sequencing Data
- Excellent webcast who prefer to sit back and watch: Fine-tuning CNV Analysis for the Clinical Analysis of NGS Samples
While there are many tools for calling CNVs from NGS data, VS-CNV is unrivaled in its robust annotation, computational analysis, filtering, and clinical interpretation capabilities. If you have any questions or comments about the information presented here or about our software, feel free to enter them into the comments below or email us at [email protected].