Our customers often ask if they can include quality control metrics in their final reports. While which metrics you actually need to report may be unique to your lab, there are a variety of metrics that we can immediately render into a report, and even more that can be rendered with a few customizations.
Reporting Average Coverage QC Metrics
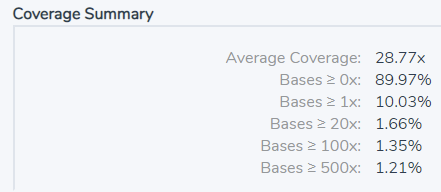
One flavor of quality control metric would be the ability to report the average coverage across the entire sample. This will require having access to the BAM or CRAM file, along with a way to define target regions, either through a BED file or by using a binned coverage approach. After coverage is run, we can include coverage summary metrics in the final report (Figure 1).
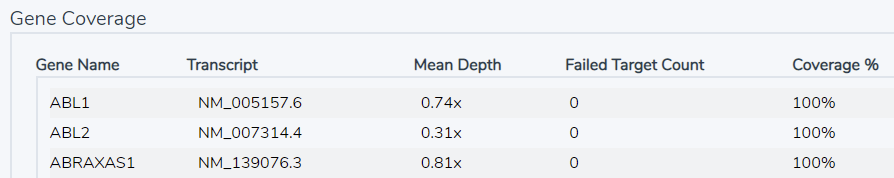
By bringing in a Gene Panel with our coverage regions, as described above, we can get more specific, reporting on the coverage and failed targets on a gene by gene basis (Figure 2).
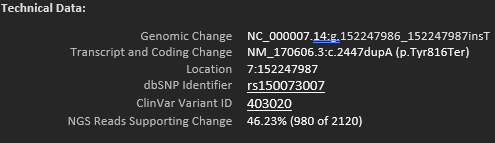
We can get more specific again, looking at the quality of specific variants. This could be the read depth, or the GQ score for a specific position, or the overall reads supporting a variant call (Figure 3). Here we have an example of a variant where out of a read depth of 2120, 980 were for the alternate call.
There are many levels of quality control metrics that can be incorporated into a final report. From using our Sample Statistics algorithm which generate information on a sample-wide basis, to our CNV quality control flags, we are broad on reporting possibilities.
Please reach out to our FAS team at [email protected] if you have specific reporting needs and we will be happy to help you!