In many cases, VarSeq users typically run single trio projects or perhaps an extended family project. Not only are all the inheritance model algorithms available in the VarSeq software to capture de novo, dominant, or recessively inherited variants but there are a number of quality control fields to help ensure the pedigree was set up properly. The last thing any user wants to find out that their family relationships were inaccurately designated since this would create many inheritance errors with downstream analysis. The best place to review the designated pedigree information and family quality control check is the samples table as seen in Figure 1.
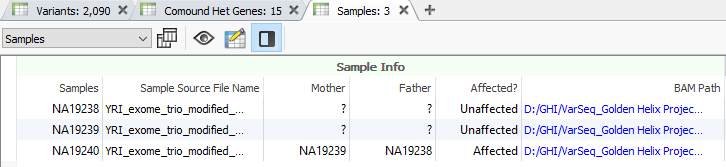
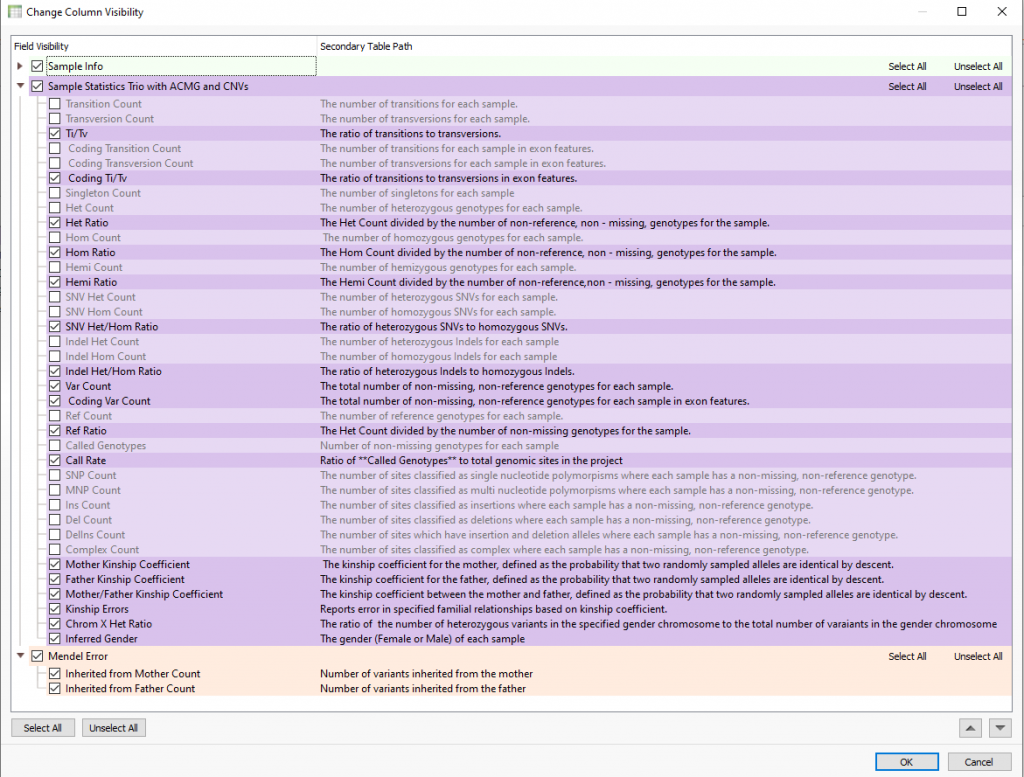
Not only is the user presented with the family relationship designated with the family import option or family manifest file (typically used for extended families), but will also explore many sample statistics. Aside from general statistics such a transition and transversion counts or heterozygous/homozygous counts for variants, the family-specific stats include mother/father kinship coefficients, kinship errors, and inferred genders (Figure2). As seen at the bottom of Figure 2, users now have a new set of fields in VarSeq 2.2.2 called Mendel error. The general application here is to ensure that the family designation is correctly set by reviewing the number of the probands’ variants inherited from mother and father. In other words, if the samples were not appropriately labeled, the number of “inherited from” variants would be either too low or missing and not make sense. You can see examples of these “inherited from” counts in Figure 3, where the proband has approximately 64K variants inherited from mother and 68K from father.
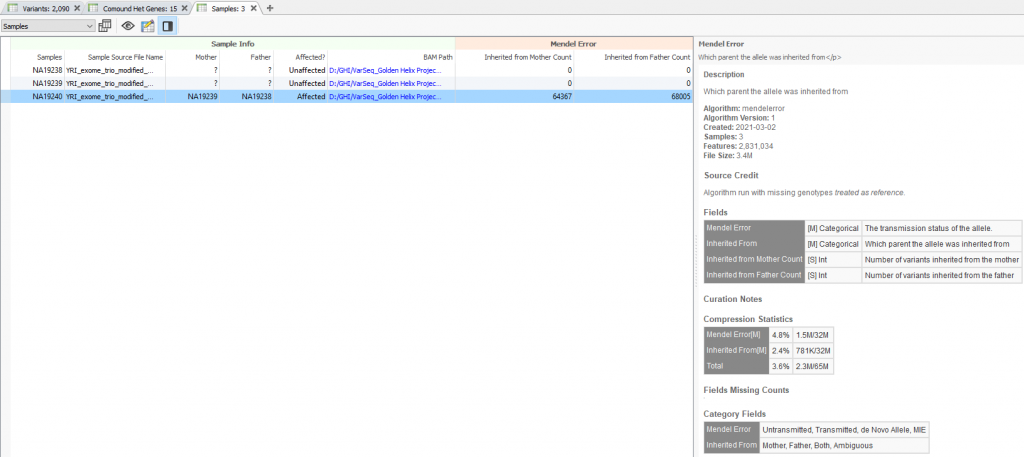
Despite this being a simple example and introduction to the tool, this new feature would be crucial to quality checking a massive multi-family project which some of our users explore in a research application. VarSeq has a huge range of potential use in both the clinical and research realm. If you would like to learn more about setting up your clinical or research next-gen sequencing project, please contact us at [email protected]. Feel free to also check out some of our other blogs that contain important, useful news and updates for the next-gen sequencing community.