In this blog, we will be covering new assessment catalogs and how they work to improve saving and tracking variant interpretations. VarSeq is a variant analysis tool that effectively analyzes single nucleotide (SNVs) and copy number variants (CNVs) in both cancer and germline workflows. Because VarSeq enables such diverse variant analysis, there are many research labs and institutions that evaluate both SNVs and CNVs not only as they pertain to cancer, but also their impact on germline diseases. As more samples are collected and more variants are annotated and classified, the value of tracking and saving these variant interpretations becomes evident. To aid in this process, VarSeq uses assessment catalogs to save and organize variant interpretations. Since the release of VarSeq 2.2.2, cataloging variants has been improved to better accommodate saving germline vs cancer variants but also SNV vs CNV classifications and interpretations. In this blog, I plan to compare the previous assessment catalogs that were available in VarSeq 2.2.1 to the updated assessment catalogs which improve the saving and tracking of variants.
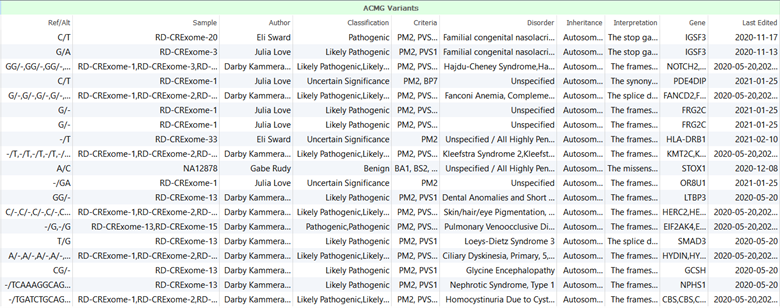
In general, VarSeq assessment catalogs are used to save key information that is collected for variants which can ultimately be used to generate clinical reports. As variant interpretations and classifications are saved, if the variant were encountered again in another sample, the interpretation would automatically fill in saving the user time and effort. Assessment catalogs save a lot of information for variants including but not limited to variant information, which user evaluated the variant, which ACMG/AMP criteria were applied to classify the variant, and the cancer type or disorder for which the interpretation was collected. Figure 1 shows an example of an assessment catalog contents.
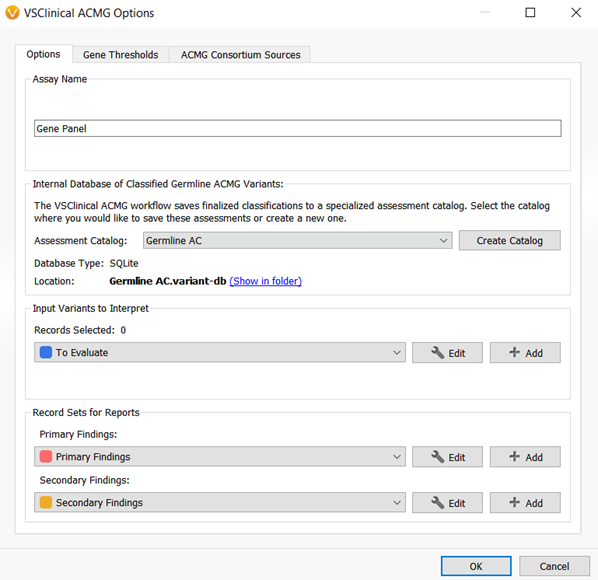
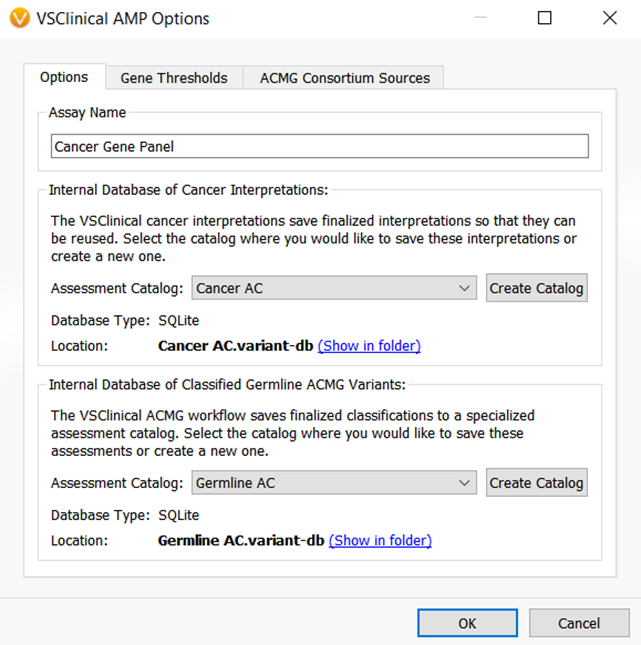
The VarSeq 2.2.1 assessment catalogs could save single nucleotide variant classifications and interpretations for both cancer and germline workflows. Figures 2 and 3 show the VarSeq 2.2.1 assessment catalogs that are available for germline and cancer variant analysis, respectively.
However, what if your lab is analyzing CNVs? What if you want to save and catalog notes from the pathology lab on certain variants? Or, what if you want to save variant level interpretations for somatic variants as well as biomarker level cancer interpretations? These questions inspired the need to update and create new VarSeq assessment catalogs to include more information in the interpretation, and better accommodate those users that analyze SNVs and CNVs in both cancer and germline spaces.
The updated assessment catalogs currently available in VarSeq 2.2.2 address the needs mentioned above and then some! Assessment catalogs now include more fields that draw information from annotations such as OMIM and MONDO IDs, can save notes on the interpretations or comments on applied criteria, and will record the citations that were referenced within the evaluation.
With the addition of the ability to evaluate and classify CNVs according to the ACMG guidelines within VSClinical, two new assessment catalogs were created. One assessment catalog will save sample level interpretations and classifications for CNVs, and another catalog will capture the CNV interpretations at a gene level. Capturing CNV interpretations at both the CNV and gene-level is important for CNV workflows as the information on certain genes or CNVs can be limited. The assessment catalogs allow your lab to essentially build an internal database for interpretations and classifications for the CNVs and genes that you evaluate within VSClinical (Figure 4).
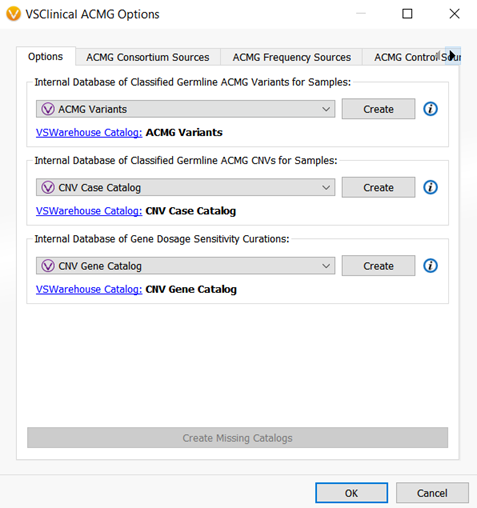
There is one more catalog that was added to VSClinical and that is the Somatic Catalog. This catalog is the AMP workflow equivalent of the Classified Germline ACMG catalog and will record oncogenicity scoring interpretations and classification for a sample. (Figure 5).
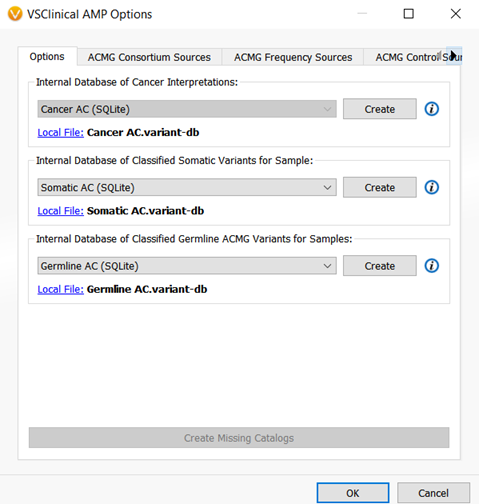
The new assessment catalogs allow users to save, organize, and track variants more effectively ultimately expanding the application’s saved variant interpretations for generating clinical reports and for annotating new projects. If you have not yet switched over to using the new assessment catalogs, I would encourage you to do so! You do not need to worry about losing existing variant interpretations as any existing assessment catalogs can be easily converted so that future variants that are saved to the catalog can have full capability with the added fields.
If you have any questions about getting set up with the new and improved assessment catalogs, tracking variant interpretations, or how you can further utilize assessment catalogs in your VarSeq projects, please do not hesitate to reach out to our technical support team at [email protected]. Feel free to also check out some of our other blogs that contain important, useful news and updates for the next-gen sequencing community. Thanks for reading!