Today I ran into an interesting fact about how a prolifically used catalog of population controls classifies African Americans with potential impacts on research outcomes. The 1000 Genomes Project is arguably our best common set of controls used in genomic studies. They recently finished what was termed as “Phase 1” of the project, and they have been releasing full sets… Read more »
Bioinformatics is a serious thing. Petabytes of data that hold the truth to the genetic underpinnings of the human species, among many others, are generated and analyzed by the world’s leading scientists every year. Here at Golden Helix, we thought that there was plenty of room to add a little humor into the bioinformatics world. Meet Dr. X. He is… Read more »
Type 2 Diabetes, Rheumatoid Arthritis, Obesity, Chrohn’s Diseases and Coronary Heart Disease are examples of common, chronic diseases that have a significant genetic component. It should be no surprise that these diseases have been the target of much genetic research. Yet over the past decade, the tools of our research efforts have failed to unravel the complete biological architecture of… Read more »
Editor’s Note: This case study was written while Dr. Gonzalo Rincon was with the University of California, Davis. Dr. Rincon is now working as a Principal Investigator in Animal Genetics at Zoetis. Gonzalo Rincon, DVM is a Project Scientist in the Medrano Lab, part of the Department of Animal Science, at the University of California, Davis. While the lab works… Read more »
My work in the GHI analytical services department gives me the opportunity to handle data from a variety of sources. I have learned over time that every genotyping platform has its own personality. Every time we get data from a new chip, I tend to learn something new about the quirks of genotyping technology. I usually discover these quirks the… Read more »
The prevalence of open-source bioinformatic tools in the genetic research space is enormous. According to The North Shore LIJ Research Institute, there are over 500 genetic analysis software packages – the great majority of which are free – as of August 2010. Open-source tools are incredibly important in genetics. They allow new methodologies to be created and expanded. They tie… Read more »
I’m a huge supporter of the Free and Open Source Software movement. I’ve written more about R than anything else on my blog, all the code I post on my blog is free and open-source, and a while back I invited you to steal my blog under a cc-by-sa license. Every now and then, however, something comes along that just might be worth paying… Read more »
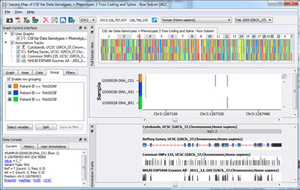
Recently several customers have asked how SNP & Variation Suite (SVS) treats gender when calculating genotype statistics. In this blog post, I will cover SVS’ current capabilities, what we have available through Python scripts, and what is coming in the near future. We thank all of our customers who have inquired about these capabilities and have given us valuable feedback… Read more »
Dr. Julia Pinsonneault is a Research Scientist at The Ohio State University where she works to find biomarkers that guide effective treatment. Published recently in Neuropsychopharmacology, Julia has found success using SVS to find novel associations that move her research forward. Usually managing 3-5 research projects at a time from various cohorts, she found time in her busy schedule this… Read more »
As most in the Golden Helix community are aware, SNP & Variation Suite (SVS) can handle all sorts of data including files from Affymetrix, Illumina, Agilent, and Complete Genomics with its powerful data management capabilities. Announced in February, SVS is now part of the Pacific Biosciences Partner Program and has the ability to analyze PacBio files.
This month Biostatistics published online an open access article I co-authored with Dr. Laura Black from Montana State University: “Learning From Our GWAS Mistakes: From Experimental Design To Scientific Method.” The paper version is expected to come out in the April 2012 issue. I’m hoping that you will take the time to read it. And I’m hoping you will violently… Read more »
In the last couple of weeks, the SVS Script Repository has seen a handful of new additions. This blog post highlights three new scripts, but as always, we welcome you to check out our repository regularly to enjoy the new and exciting functionality made possible by our Python integration in SVS! (To get these, or any other scripts, simply go… Read more »
It’s that time again! We here at Golden Helix are excited to announce SVS 7.6 with more features for DNA-Seq analysis, the addition of RNA-Seq functionality, reorganization of SVS into “packages” including two new ones, and the release of new plot types enabled by Matplotlib. It’s certainly been busy as we pack all this into the sixth installment of the… Read more »
Allow me to introduce you to Blaine Bettinger. Blaine is a patent attorney who holds a PhD in Biochemistry with a concentration in genetics. He is also a family history enthusiast who writes the Genetic Genealogist blog, where he gives commentary on applications of genomic science for advancing personal and family history research. I first learned about Blaine last May… Read more »
The turning of the calendar from 2011 to 2012 has been a good time for me to reflect on the lessons of the year and make resolutions for the new one. It is also the opportunity to step back and look at some of the larger systemic trends in our field and think about whether we are doing as much… Read more »
Dr. Raman Babu is a Maize Molecular Breeder at the International Maize and Wheat Improvement Center (CIMMYT). Like his counterparts conducting human genetic research, Babu used to rely entirely on free, open-source tools to complete his work. Frustrated with continual crashes and technology that was developed in the pre-SNP era, Babu switched to SNP & Variation Suite (SVS) almost a… Read more »
Recently we have expanded our annotation track offerings with new human variant frequency catalogs such as the 1000 Genomes Phase 1 Data. Of course, we also curate data for plant and animal genomes – some of which are currently available in our software and some of which will be available in our next release. In this blog post, I will… Read more »
Have you ever chosen to do something yourself instead of paying money for the alternative, when, in the long run, the alternative is not really that expensive? We all have, as we all have limited budgets. One particular example comes to mind in my own life. My boyfriend, Ben, and I bought a house in April. Unfortunately, it wasn’t properly… Read more »
While there is a lot of excitement in the industry about Next-Generation Sequencing, questions remain as to whether it will be “the answer” to the ails of the genetic research industry. I recently had the opportunity to sit down with three of Golden Helix’s thought leaders and ask a few questions about NGS – Gabe Rudy, Vice President of Product… Read more »
If you have ever worked with NGS variant data, you may have come to realize that the first task at hand is the seemingly simple categorization of your variants into two bins: known and novel. Of course, if you’ve ever worked with NGS variant data, you may have also come to the realization that this step is more complex than… Read more »