Our FAS team would like to thank everyone who attended our December 2022 webcast, A User’s Perspective: Somatic Variant Analysis in VarSeq 2.3.0. This webcast allowed three members of our FAS team to give their unique insights concerning the improvements to our new VarSeq 2.3.0 release, which will be recapped here. Starting with template creation, our Technical Field Application Scientist,… Read more »
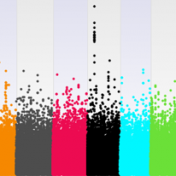
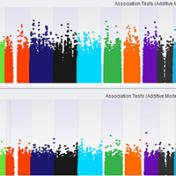
SVS offers several options to conduct genome wide association tests and mixed linear models. At times, it can be challenging to decide which test, model, or adjustments to use when setting up your analysis. I want to briefly explore the options available in SVS for association tests, and mixed linear models to hopefully facilitate in understanding and choosing which options… Read more »
SVS is a project-oriented program that manages and analyzes genomic datasets. This webcast statistically and visually explores the relationships among genetic variants within a cattle dataset. Even further, this webcast evaluates genotypes with corresponding phenotypes to assess how well a model can predict a phenotype of interest. Starting with genotypic data from the microarray and the recorded phenotypic data for… Read more »
As our final part of the ‘Top-Quality GWAS Analysis’ blog series, we will be giving a summary of the values behind GWAS quality control and quality assessment. Performing GWAS can provide insight into the association of genetic variants with traits and complex disorders. Any novel insights into marker-phenotype associations need to be based on performing quality control steps. In this… Read more »
Population Stratification This article is going to cover how to factor for population stratification in your association test to continue our blog series on top quality GWAS analysis (additional articles for this series are located at the bottom of this blog). Quality control steps up to this point have included assessing sample and marker statistics, LD pruning on markers, and… Read more »
Sample Relatedness Pruning your data based on Linkage Disequilibrium (LD) values and filtering for sample “relatedness” are ideal quality assurance steps following the marker and sample quality filtering described in Part II of this blog series. The value of running an Identity by Decent estimation not only allows you to factor family relatedness in your samples but makes screening for… Read more »
Eliminate Low-Quality Samples and Markers In Part I of this GWAS Analysis series, Dr. Eli Sward provided us with a great overview on the value SVS provides in managing the quality of your SNP or NGS data to maintain the high power and accuracy of your GWAS. He also gave a snapshot of what a typical genotype spreadsheet may look… Read more »
Importance of Quality in Association Tests SVS is a research application platform provided by Golden Helix that enables an array of computational analyses including genome-wide association studies (GWAS). GWAS is an observational study that can provide insight into the association of genetic variants with traits and complex disorders. The foundation of GWAS utilizes large cohorts sequenced with single nucleotide polymorphisms… Read more »
Genome-wide association studies (GWAS) are useful in genetics as they test for the association of a phenotype with common genetic variants. GWAS is “hypothesis-free” and does not require prior knowledge of a gene’s biological impact on a trait. The catch though is that this leads to analyzing hundreds to thousands of genome-wide array samples to elucidate single nucleotide polymorphisms (SNPs) associated with a specific phenotype.
In case you missed our live event yesterday, I wanted to share a link to the webcast recording: New Enhancements: GWAS Workflows with SVS. There were several questions asked, so we’ve also shared the Q & A session below! Question: Are these enhancements priced as a separate feature? Answer: No, SVS is a constantly evolving platform, so everything you see in this webcast is… Read more »
When studying complex diseases that may have multi-genic contributions from across the genome, it is not uncommon to see what may appear like elevated correlation between your trait or other test variable and the SNPs across the genome. The problem is at first glance you won’t be able to tell if this is due to a population structure in your… Read more »
Genome-wide association study (GWAS) technology has been a primary method for identifying the genes responsible for diseases and other traits for the past ten years. GWAS continues to be highly relevant as a scientific method. Over 2000 human GWAS reports now appear in scientific journals. In fact, we see its adoption increasing beyond the human-centric research into the world of… Read more »
ASHG 2016 is in our rear mirror. Again, it was bigger and better than the previous year. The conference hosted over 9,000 visitors from 66 countries. This gave the event a level of vibrancy that was evenly matched by the wonderful ambiance of the city of Vancouver. Nestled in between the two conference centers was a little pier offering spectacular… Read more »
Dr. Folefac Aminkeng, first place winner in this year’s Abstract Challenge, will present his work to the community via a live webcast on Wednesday, April 20th at noon EDT. Dr. Aminkeng is a Postdoctoral Fellow at The Centre for Molecular Medicine and Therapeutics (CMMT) at the University of British Columbia in Vancouver, BC. Aminkeng’s research focus is the pharmacogenomics of adverse… Read more »
Customer success is very important to us at Golden Helix. Every month we showcase their success by putting together a blog post highlighting the most recent publications. We have been compiling this list since 2003 (you can find the full list here). Today, I am very honored to announce that the Golden Helix software has assisted in one thousand publications. We have enjoyed… Read more »
Concepts and Relevance of Genome-Wide Association Studies Genome-Wide Association Studies continue to be a very effective method for determining the underlying cause of disease, Golden Helix is happy to share our long-standing knowledge with the community. We are very happy to announce that “Concepts and Relevance of Genome-Wide Association Studies”, a paper surrounding GWAS was recently published in Science Progress… Read more »
Thank you to all who submitted an abstract in the 2016 Abstract Challenge, it was a great success! With more than 30 submissions and a wide variety of topics, selecting only 3 winners very difficult! With that being said, here are the top 3 winners for this year’s Abstract Challenge: Our 1st place winner is Dr. Folefac Aminkeng, Postdoctoral Fellow… Read more »
With the release of our updated GWAS E-book, we have recently updated the GWAS example project (SNP Genome-Wide Association Tutorial – Complete). This updated project includes more details about how spreadsheets were generated, how to generate plots and which images were used for the GWAS E-book. This information can be found in the User Notes view in the project navigator and… Read more »
Recent Customer Publications We hope everyone had a Merry Christmas! The Golden Helix team is looking forward to the start of a new year, but first we wanted to share our last round of customer publications from 2015. Here’s to many more in 2016! Bishwa Sapkota and Dharambir Sanghera of the Oklahoma University Health Sciences Center and colleagues published The Genome-wide Association Study of… Read more »
Give our SVS viewer a try today! Interested in seeing what the SNP & Variation Suite (SVS) software can do? Download the free SVS Viewer! With the SVS Viewer, you can explore and interact with the workflows of a pre-built projects. To get you started, we have included a SNP GWAS project for you to download. And don’t worry, it is… Read more »