Since we released our Phenotype Gene Ranking algorithm in VarSeq, it has become a staple of the way people conduct their analysis. It allows for a combination of filtering with ranking to prioritize follow-up interpretations of analysis results.
Our PhoRank algorithm will be available in our upcoming SVS release to also aid in the numerous research workflows performed on SNPs and variants.
Rank Genotypes, Association Test Results or Anything In-Between
The PhoRank algorithm ultimately gives you the ability to associate phenotype terms from the Human Phenotype Ontology to the genes in your dataset.
It may be useful to see how your genomic data is linked to specific phenotype terms at various points of the analysis process:
- After using a variant annotation algorithm to filter variants in your dataset down to certain functional and population frequency criteria
- After doing some genotype filtering based on Marker Statistics such as your cohort allele frequency
- After doing association tests such as in a standard GWAS workflow and sub-setting down to candidate SNPs
If your SNP or variant spreadsheets in SVS have a marker map applied, you will be able to launch the new PhoRank dialog:
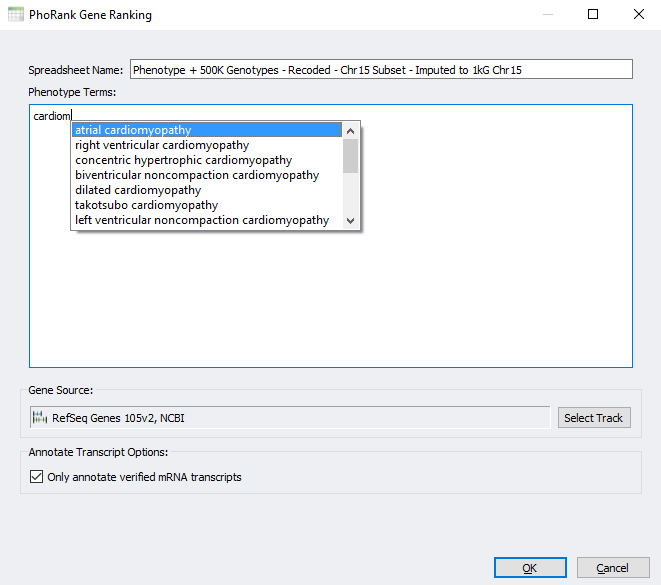
From here you can type in phenotype terms, and valid HPO terms that contain your input will be provided as potential completions.
We also detect the current species and genome assembly of the project. We then select the default gene track to use for mapping the marker positions in your spreadsheet to a list of genes to rank. You can also select other gene sources of your choosing.
Note that since the Human Phenotype Ontology is about mapping human phenotypes to human gene names, it may not be that useful to use this feature outside of human genetics. Although with model organisms that share many genes (with the same HUGO gene names), it would be completely valid to explore the results of PhoRank and see how much it makes sense for your research goals.
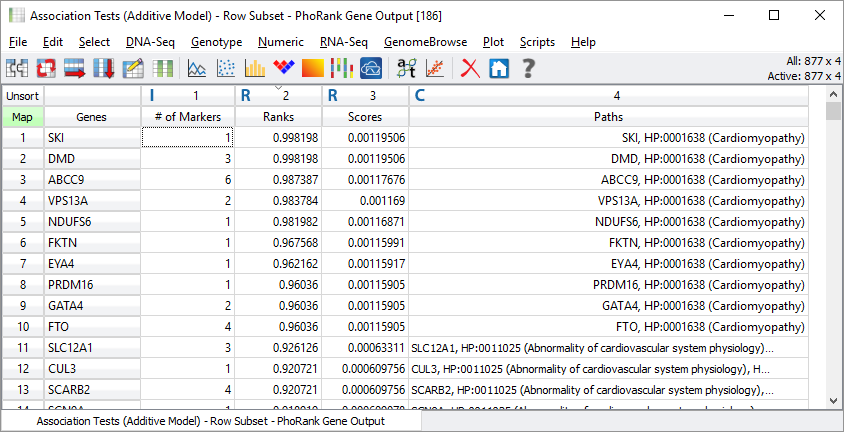
After the algorithm runs, SVS provides the list of genes and their PhoRank rank statistics. For each gene, we summarize how many of the markers in the input spreadsheet are contained within that gene. If you sort by the Ranks column, you get a quick sense of which are the genes with the closest association with the input terms. The Path column provides the evidence of the most direct path from the input terms, through the HPO and GO ontologies, and to the target gene.
We are always excited to expand the capabilities of our flagship research product SNP and Variation Suite, and with the addition of PhoRank we are bringing another popular innovation to the feature-rich platform.