Explore the cutting-edge capabilities of VarSeq in prenatal genetic screening as we delve into real-life cases, expert analysis, and efficient strategies to quickly assess Whole Exome Sequencing (WES) samples for genetic abnormalities in our recent webinar.
Thank you all to those who attended our VarSeq webinar covering prenatal genetic screening! We had a great turnout and loved hearing from our customers in the webinar comments section. This webinar is a companion broadcast to our latest eBook, written by the Golden Helix CEO, Dr. Andreas Scherer. This eBook, Prenatal Genetics, has been downloaded hundreds of times in the few days since its release. Covering the current state of prenatal genetic testing, it also gives several examples of how VarSeq can be used to quickly assess prenatal WES samples for genetic abnormalities. These abnormalities range in size from a chromosomal trisomy event to several small single variant changes. In our webcast, our FAS team, with Dr. Jennifer Dankoff and Solomon Reinman, our Technical FAS, bring these variants to life, demonstrating applicable filtering strategies and taking us all the way to rendering final clinical reports.
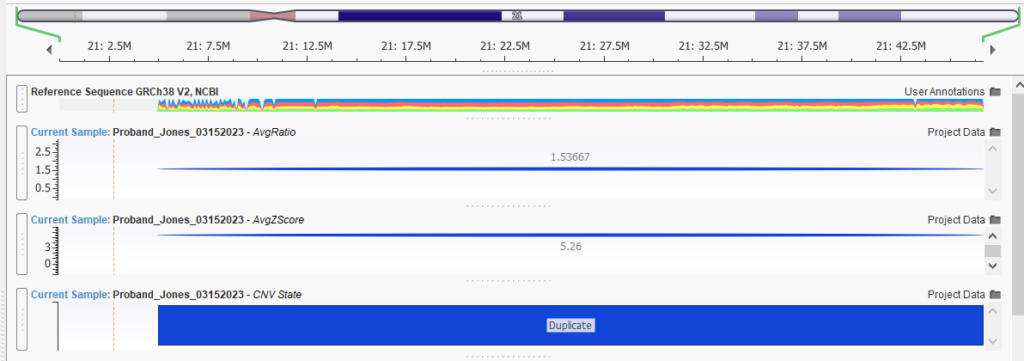
Our webinar started off with Jennifer demonstrating Trisomy 21 detection from a WES sample. Consistently high coverage across chromosome 21 resulted in a ratio of over 1.5, as seen in Figure 1. Coupled with the strong Z-score, there was sufficient evidence to call a duplication across chromosome 21.
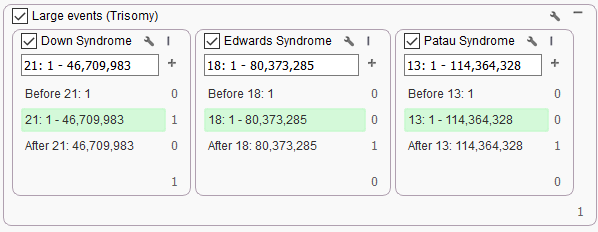
Although this project had a specific chromosome 21 duplication, the filter chain used to isolate that CNV call was broad enough to identify other large or chromosomal level (Figure 2). Here we are able to identify CNVs in chromosomes 18 and 13 for Edwards Syndrome and Patau Syndrome, respectively. We are not limited to these CNVs, and could also set up unique filter chains for isolating smaller events or chromosome-wide events affecting the sex chromosomes. Through a quick identification, we can move right into analysis with VSClinical to render the final report that can get back to the family.
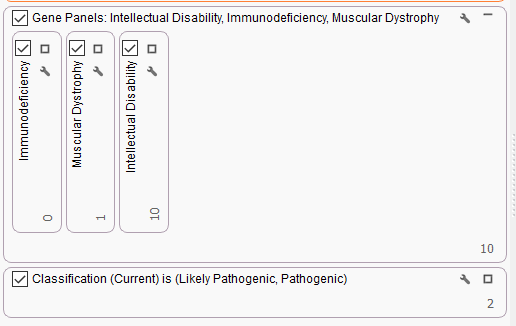
Analysis with VarSeq and VSClinical is not limited to these large CNV events. Through WES sequencing and analysis, we also have several examples of monogenic diseases identified through VarSeq analysis. Solomon first took us through an example of a male proband who has a family history of Duchenne Muscular Dystrophy. Here, Solomon takes us through a workflow that has been specifically customized to search for pathogenic variants associated with DMD (Figure 3). To do this, there are three gene panels put into a parallel filtering strategy. These panels include genes associated with immunodeficiency, muscular dystrophy, and intellectual disability. By putting these panels in parallel, we can see which variants fall through a specific panel. Last, the ACMG auto-classifier is used to isolate two pathogenic variants for this proband.
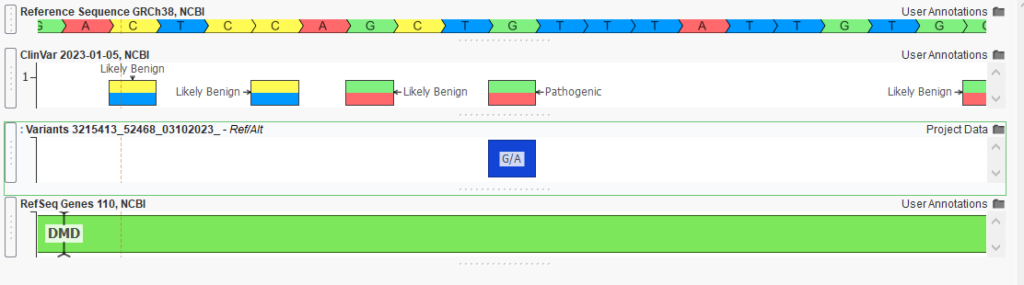
One of these pathogenic variants is the DMD G to A change, resulting in a stop gain for the gene (Figure 4). By quickly isolating these variants, we can bring this information into VSClinical, where the ACMG classifier contributes pathogenicity information to what will be our final clinical report. The clinician is then able to render the report and return this information to the waiting family.
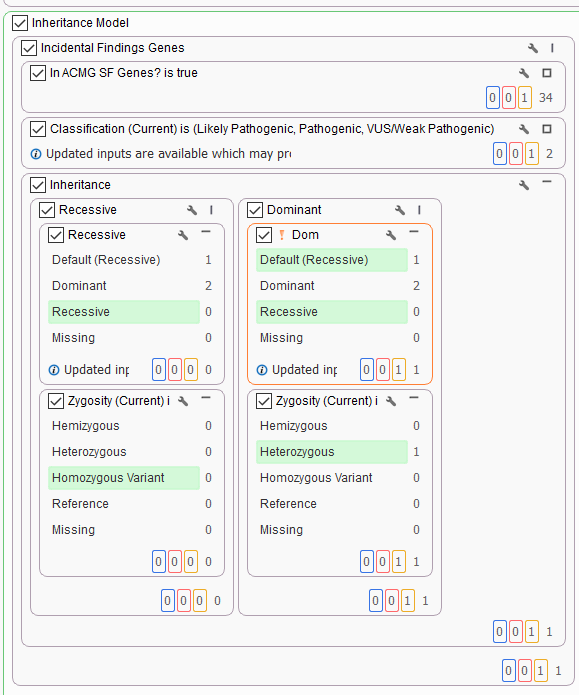
For our final project, Solomon had an example with a trio where an unexpected secondary finding is isolated. This sample in question was brought in for routine WES screening. While there are a number of de novo variants and dominant inherited variants isolated using this workflow, we also see the benefit of having a secondary findings column (Figure 5). The inheritance filter has isolated a dominant heterozygous variant that is in a gene specified by the ACMG Secondary Findings gene panel.
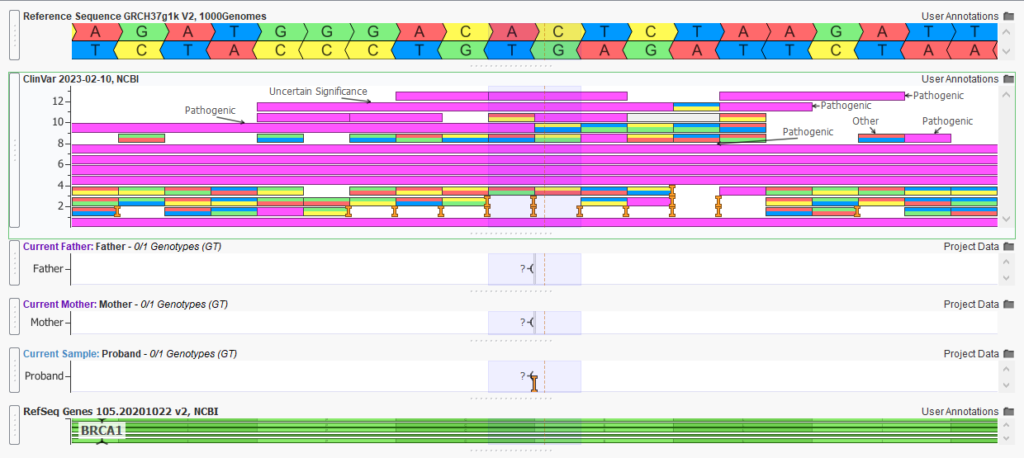
The variant of interest, in this case, is a pathogenic de novo frameshift variant (Figure 6). By using our VarSeq workflow to isolate this variant, we have the flexibility to identify and report on secondary findings. The clinician is able to decide if this information should be reported to the family, as mutations in BRCA1 associated with hereditary breast and ovarian cancer susceptibility. Persons with a family history of certain types of cancer or who are found to carry a genetic mutation associated with hereditary cancer may be advised to undergo more frequent or earlier cancer screening, consider preventive surgeries or take other risk-reducing measures. With VarSeq and VSClinical, we have the option of isolating and reporting this and other secondary findings to the clinicians.
When adopting WES prenatal screening, there are a number of unique challenges that may arise. For example, a small clinic may be concerned with interpreting vast amounts of data in a timely manner or increased cost per samples. With routine analysis through VarSeq and VSClinical, we offer the ability to streamline project creation and analysis through customizable project templates. The main value of this is the ability to minimize the turnaround time from sample analysis to the point where the clinician delivers results to families. VarSeq gives the option to isolate and report on secondary findings. The Golden Helix model charges per licensed user, not per sample, so labs are able to quickly meet new load size challenges. In addition, with advances in WES and analysis, the resolution for detecting genetic abnormalities increases, and we are able to provide quality reporting to families.
And with that, we had some audience questions for this webcast:
1.Can you detect a single exon deletion or duplication in the DMD gene?
Yes, with VarSeq CNV calling, we are able to detect copy number variants from whole chromosome level events down to one exon in size. For more on using CNV calling with VarSeq, please see our webcast on Handling a Variety of CNV Caller Inputs with VarSeq.
2. Can Triploidies be detected with VarSeq technologies?
Absolutely! We are able to detect duplications, heterozygous deletions, full deletions, and even LOH using our coverage regions algorithm.
3. Is VarSeq restricted to these diseases, or can it be used for other diseases: Gaucher or Fabry disease?
Yes, we can certainly build workflows to specifically isolate mutations causative of those diseases, or we could build broader workflows to isolate recessive markers, dominant markers, or X-linked diseases. This webcast on Efficient Application of NGS Family-Based Analysis goes into more detail on family screening and may be of interest to you.
4. Is VarSeq only a prenatal analysis platform? Can it be used for other use cases?
Good question! VarSeq can be used for a wide variety of use cases. For example, and not limited to, we support trio analysis, single sample analysis, cohort analysis, CNV calling and analysis, rare variant and phenotypic association analysis, and a variety of cancer analysis. Feel free to contact us and let us know more about your specific needs and how we can help develop custom solutions.
Thank you all again to those who came to participate in our Prenatal Genetics Screening with VarSeq webcast. We had a lot of fun highlighting this use case for VarSeq in the rapidly growing field of prenatal NGS testing. Our Golden Helix customers have unlimited access to FAS training sessions, so let us know how we can help you implement prenatal NGS testing in your lab. If you have any questions about how VarSeq could suit your unique use case, please reach out to [email protected], and we will be happy to set up a demonstration!