In the last year, we have seen a surge in customers moving to whole genome sequencing. Not only does whole genome sequencing provide unparalleled gene coverage compared to whole exome, but depending on the kind of sequencing, you may expect to see additional file types as well. For example, our partners at PacBio will provide an additional VCF containing copy number variants and structural variant calls. If you want to learn more about our partnerships with PacBio and Twist, please watch this webcast here.
Importing and Linking CNVs and Structural Variants in VarSeq
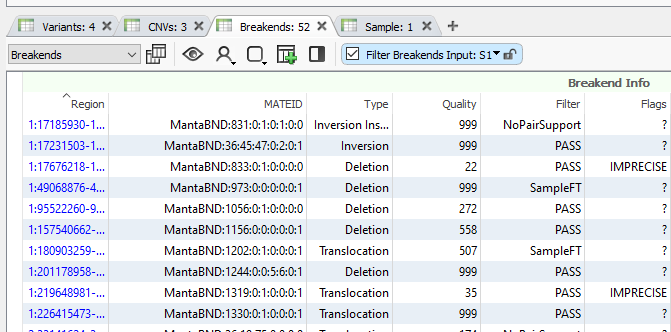
Moving away from small variants (your typical SNPs, INDELs, etc.) and into copy number variants and structural variants, the format of the data becomes, understandably, much more complicated. For example, when looking at a single fusion or breakend structural variant event, you will two entries in your VCF. The first entry will contain the first MATEID, and the second, somewhere else in the VCF, will show the partner MATEID. The trick when importing all the variants, copy number variants, and structural variants into VarSeq is not only to designate the appropriate tables for each of these variant types, but also link together the complicated MATEID structures for easy annotation and analysis.
Streamlining Complex Variant Imports with VarSeq-CNV
Luckily, VarSeq makes this process easy, like Sunday morning. With our VarSeq-CNV module, we have our comprehensive data importer. This data importer will automatically parse out the small variants, copy number variants, and structural variants (or Breakends) into their own unique tables (Figure 1). Not only that, but the complicated MATEIDs will be automatically paired, making annotation and analysis a breeze.
To use the importer, simply confirm that your structural variants VCF is in a standard VCF format and that you have the VarSeq-CNV module turned on in your license. For our current customers, our FAS team is happy to help set up your workflow through a series of one-on-one training sessions.
If you want to discuss how VarSeq can improve your analysis, please contact us at [email protected] or fill out an eval request here.