We are excited to announce the release of our gnomAD v4 annotation tracks for VarSeq. This version of the GnomAD database represents a significant leap forward, including data from an impressive cohort of over 800,000 individuals — a remarkable 5x expansion compared to the previous releases. Notably, this dataset is comprised of two distinct callsets: exome sequencing data from 730,947 individuals, including a substantial contribution from the UK Biobank, and genome sequencing data from 76,215 individuals.
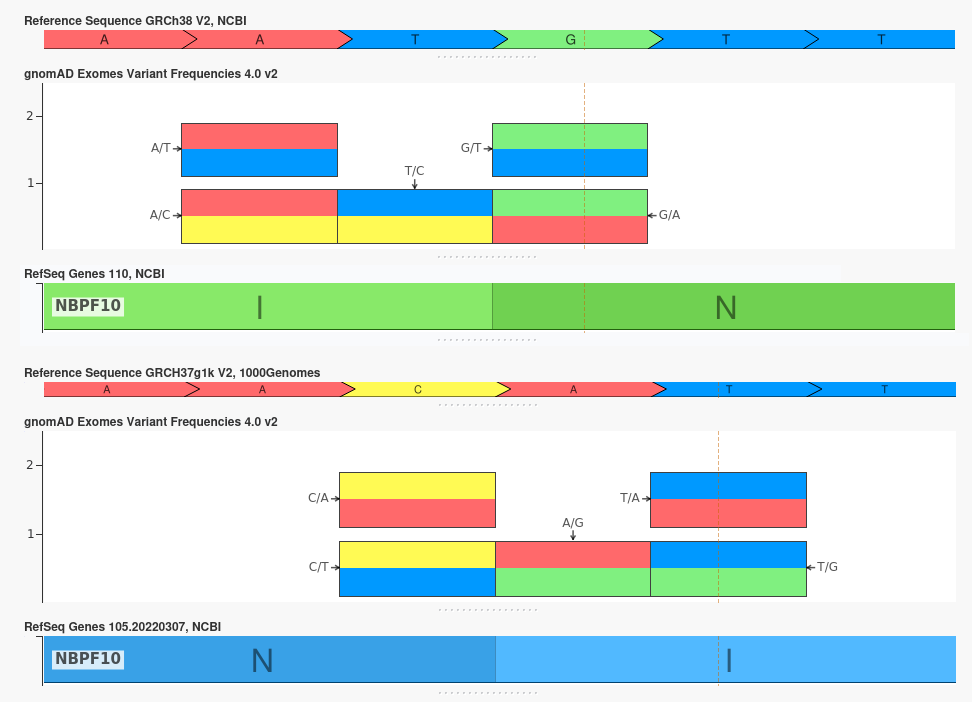
However, because the variants in gnomAD v4 have been aligned exclusively to the GRCh38 reference assembly, additional work is required to curate a GRCh37 version of the new tracks. We converted the variant coordinates in these annotation tracks from GRCh38 to GRCh37 using the Liftover algorithm built into VarSeq. Due to differences between the GRCh37 and GRCh38 reference assemblies, the Liftover process occasionally results in a different reference allele for a given variant. This often occurs because the entire region has been inverted (i.e. reverse complemented) in the new reference assembly. When this is the case, the reference and alternates can be updated, but no update to the counts or allele frequencies associated with the variant is required.
An excellent example of this can be seen with the gene NBPF10. This was a forward-stranded gene in the GRCh37 assembly, but the surrounding region was inverted in the GRCh38 assembly, changing it to a reverse-stranded gene. As a result, the variants in this gene all have reverse-complimented Ref/Alt values, but the allele frequencies remain unchanged.
However, there are rare instances in which an update to the reference allele at a given location is not the result of an inverted region but is instead the result of a single updated reference base. In these cases, the counts and frequencies must be updated to reflect the fact that the reference allele for the variant in GRCh38 is the alternate allele in GRCh37.
When curating the GRCh37 GnomAD tracks, we update to the counts and frequencies for a given variant if the following conditions are met:
- The reference allele differs between GRCh37 and GRCh38 AND
- The reference alleles are not complementary OR
- The reference alleles are complementary, but neither of the adjacent reference bases are complementary between GRCh37 and GRCh38.
While the above heuristic correctly determines whether the counts and frequencies require an update for most variants, there are edge cases in which it fails. These edge cases occur when two or more adjacent bases are reverse complemented between GRCh37 and GRCh38 due to an update to those bases in isolation, rather than an inversion of the larger surrounding region. Although these edge cases are extremely rare, they will result in variant allele frequencies that are not correctly updated for a small number of variants.
Following this procedure, we are able to create a GRCh37 version of the gnomAD v4 tracks that can be used for annotation and filtering with confidence. No lifted over track is as perfect as one generated from natively aligned data, but the curated version we provide represents the best possible representation of the GRCh38 data on the GRCh37 genome.
If you have any questions about GnomAD or any other annotations available in VarSeq, please reach out to us at [email protected].