The recent release of VSClinical gives users the ability to evaluate variants based on the 33 criteria according to the American College of Medical Genetic and Genomics (ACMG) guidelines. This feature leverages a variety of variant sequencing evidence including population data, functional data, and computational predictions while providing rich visualizations and auto recommendations to help answer challenging criteria. This highly sophisticated yet user-friendly technology is intuitive, but like any new software, there is a learning curve. Our goal is to assist GoldenHelix users in becoming familiar with this new tool. This blog post will be dedicated to some fundamental aspects and techniques that can be implemented to help optimize VSClinical analysis, evaluation, and reporting of genetic variants.
Reference Genome Assembly and the ACMG Classifier
GRCh37 (hg19, g1k) and GRCh38: VSClinical is a licensed feature within VarSeq, which is designed for tertiary analysis of complex variants for gene panels, exomes, and whole genomes. This software imports sequencing data from multiple platforms with the requirements of VCF and optional BAM files, which can be converted from FASTQ files using our secondary pipeline partnership, Sentieon. When importing VCF files into VarSeq, it is required to choose a reference genome assembly for your project.
There are a few assemblies to choose from when starting your project, but not all are applicable to VSClinical. Currently, our new ACMG Guidelines interface is tested/developed for the GRCh37 g1k and GRCh38 assemblies. Some users may have projects that are built on GRCh37 hg19 and will find that they are unable to access the ACMG Guidelines in VSClinical. Not to worry though, you can build your project using the g1k assembly instead! One major difference between g1k and hg19 assemblies is the mitochondrial region. Hg19 is based on the UCSC alignment and denotes the mitochondrial region as chromosome M whereas g1k uses the rCRS mitochondrial sequence (AC:NC_012920) and denotes this region as chromosome MT. If analyzing mitochondrial variants in VarSeq and/or VSClinical, selecting the right reference genome assembly is key. However, if you are not concerned about any mitochondrial regions, you can use g1k instead of hg19 to gain access to all of VSClinical capabilities.
ACMG Classifier: Running the ACMG classifier algorithm is a prerequisite to running VSClinical and scores 18 criteria based on available evidence from 7 sources and is accessed by going to Add ‘Computed Data ACMG Classifier’. If the ACMG classifier has not been loaded before, this will require a few specific downloads that the classifier is dependent on. Additionally, first time exposure to the algorithm will have you also build a new ACMG assessment catalog. The assessment catalog is a database of previously classified variants evaluated using VSClinical so the user can prioritize their own classifications over the auto-classification output. The next window will have you define preferred transcripts and splice site boundaries if the user wants to change the default conditions. Once you have downloaded the required tracks and created an ACMG assessment catalog, the ACMG Classifier will populate your variant table on the far-right side. Any field within the ACMG Classifier, including the Classification and Auto-Classification fields, can then be used in the variant filter chain.
ACMG Classification and Auto-Classification: The Classification field in the ACMG Classifier can be used as a source of previous classifications and can be implemented into the variant filter chain. The Auto-Classification field leverages 18 of the 33 ACMG criteria to provide a classification for that variant based on the available evidence. Alternatively, the Classification field takes the auto-classification a step further and accounts for previous variant evaluations in VSClinical. These classifications are listed next to two additional fields showing the list of criteria and criteria descriptions. Any field within the ACMG Classifier can also be implemented into the filter chain, which can be useful to filter for variants of certain significance. The next step is to now get the filtered variants into the ACMG Guidelines tool.

Figure 2: The ACMG Classifier includes a Classification field that records previous variant evaluations.
Additionally, VarSeq and VSClinical are not autonomous and many properties in VSClinical can be used in VarSeq. For example, population data (GnomAD and 1000 Genomes) or functional prediction algorithms (SIFT and PolyPhen2) in VSClinical are uploaded into the variant table in VarSeq. When the ACMG Classifier is loaded, these sources are also pulled in automatically and can be used for annotating and filtering variants. The next step is to now get the filtered variants into the ACMG Guidelines tool.

Figure 3: Annotations used in VSClinical are automatically pulled into the variant table and can be used in the variant filter chain.
VSClinical Basics, Overview, Finalization
Basics: VSClinical can be accessed by clicking on the plus icon (shown below), where you’ll select ACMG Guidelines from the dropbox.
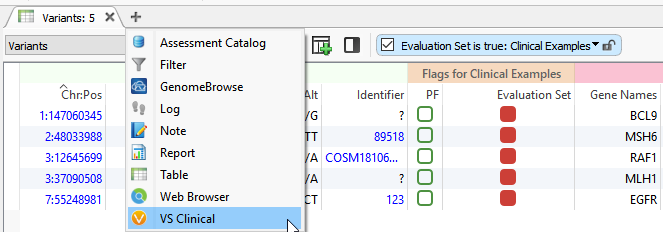
Figure 4: VSClinical can be accessed by clicking on the plus icon then selecting the ACMG Guidelines.
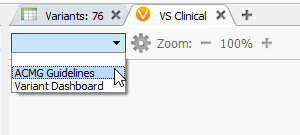
Figure 5: VSClinical can be accessed by clicking on the plus icon then selecting the ACMG Guidelines.
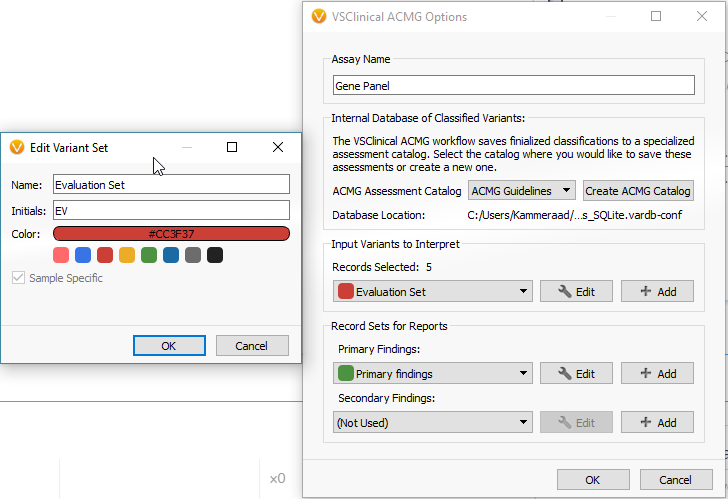
The next menu will have you select the desired assessment catalog and create the variant sets. The variant sets are a means of flagging sample specific variants to be interpreted in VSClinical. You can create a new variant set from this menu. If you are going to include your variants in a clinical report, you should also create a primary finding variant set as well.
After creating your variant sets, click OK and open the ACMG Guidelines interface showing your current sample, variants to be evaluated, and the option to start a new evaluation at the bottom. The list of filtered variants reflects the total variants meeting all the set criteria in your filter chain, and from this filtered list, you can select which variant you want to evaluate/classify (check the colored box).
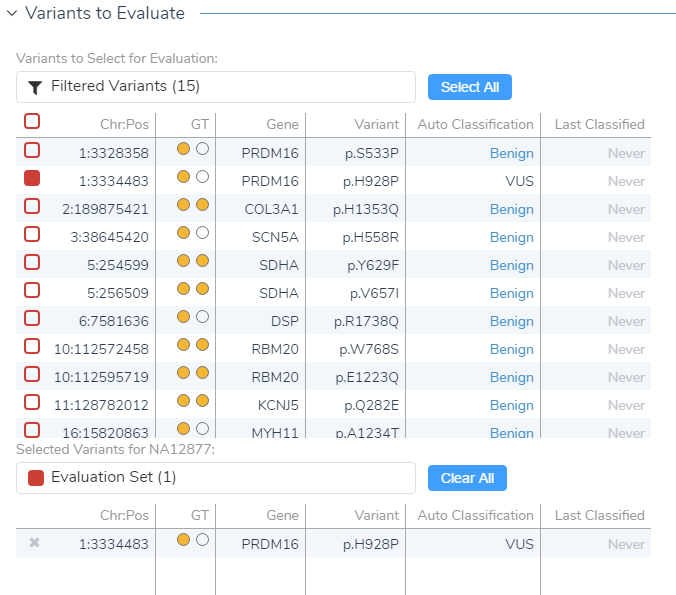
Figure 7: In VSClinical you can select the variants from your variant set that you wish to evaluate..
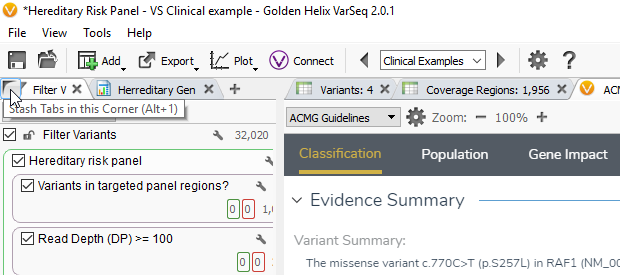
Overview: The ACMG Guidelines window is comprised of multiple tab categories including Classification, Population, Gene Impact, Studies and Clinical findings. Each tab provides specific details on the available evidence regarding that variant through which you collect the criteria for classification. If you want to have an increased visualization of VSClinical, you can hide the filter chain by hovering over the filter variants icon and clicking the corner icon or you can press Alt-1.
The Classification tab is the first window that appears in VSClinical and provides useful information regarding variant details, recommended scoring criteria and the GoldenHelix Classification prediction model. The evidence summary field provides a brief description about what is known about the variant and if the variant is present in populational catalogs or identified in ClinVar or dbSNP. The evidence summary also produces the recommended criteria to score the variant, which when answered, will populate the ACMG Criteria/Classification fields and the GoldenHelix Classifier. The GoldenHelix Classification Prediction model is an algorithm that computes a probability of each of the five classifications and offers a rich visualization for the current status of the variant. Once the evaluation is complete, it is helpful to return to the classification tab to ensure that all of the recommended criteria have been answered.

Figure 9: The Golden Helix Classification prediction model provides a rich visualization of the current classification of the variant by leveraging the answered criteria.
When answering the criteria in each section, by default, VSClinical will show only the remaining criteria left to be answered, but you can select to view all relevant criteria.

Figure 10: In each tab, this icon is displayed and can be used to show all criteria or only criteria that is relevant for the variant.
Answering criteria may not always be straightforward. To simplify this process, even more, we included not only the strength of evidence (strong, moderate, or supporting), but also ACMG reference information to the relevant criteria. After viewing the discussion for that criteria, you fundamentally choose a yes/no answer to each criteria. The user can input additional comments that can be included in the final interpretation of the variant. This representation of answering criteria is consistent through each of the tabs in the ACMG Guidelines interface (Population, Gene Impact, Studies, Clinical).
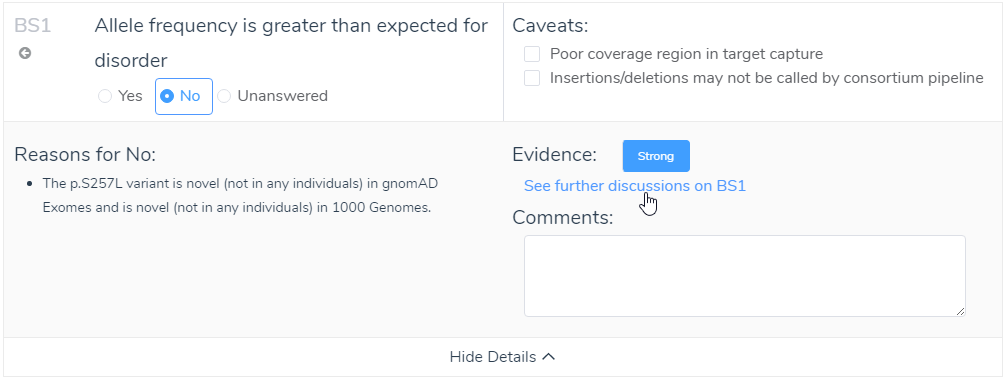
Figure 11: Utilizing available evidence in the show details for criteria can provide support for answering challenging criteria.
Finalization: After answering all the necessary criteria, and including any additional clinical references, you can report on your variant. All collected information can be pulled into the Interpretation section that is located under the Classification tab near the bottom of the page. Once the interpretation information is accepted and finalized, it will store this information in the created ACMG assessment catalog and will display the evaluated criteria in the variant table in VarSeq. If the evaluated variants in the ACMG assessment catalog are desired to be rendered in a clinical report, the flagged variants can be selected from the Primary Findings in the configuration icon (shown below).
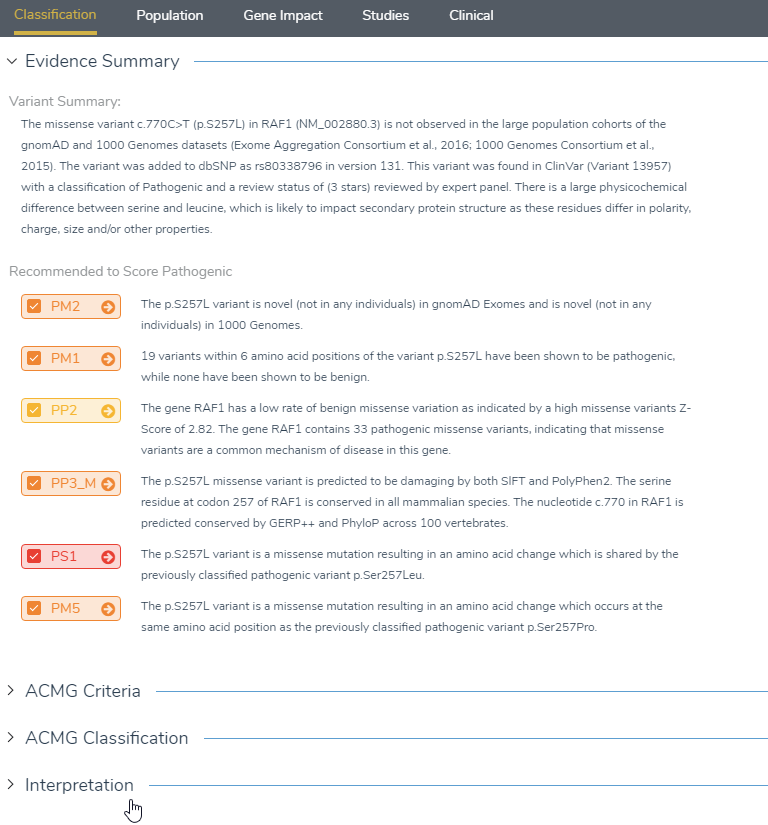
Figure 12: The classification tab will display all answered criteria under evidence summary, which can be imported into the interpretation section.
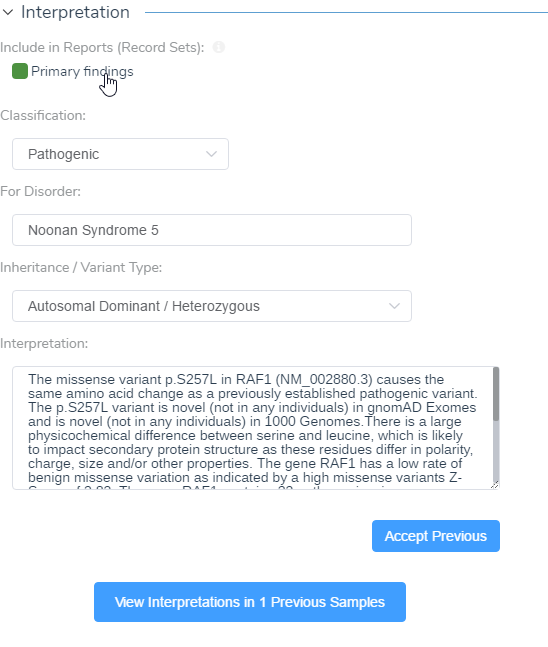
Figure 13: Primary findings can be included in the interpretation and finalizing these results will populate the ACMG Classifier in VarSeq.
Rendering a clinical report of the evaluated variants can be achieved by opening the report tab under the + icon. The report template can be changed and will include patient and sample information if available. Next, you can select the variant set under the primary findings section, will incorporate the interpretation from VSClinical. This report can then be rendered using the circle arrow icon.

Figure 15: Selecting your evaluated variant set in the primary findings will pull in the variant information and interpretation. Once validated, the chasing arrows icon at the top can be used to render the report.
Overall, there are a variety of unique functions that become more intuitive with greater time spent with the software. The goal with this blog was to assist users new to VSClinical on how to get things started. Please feel free to contact [email protected] with any additional questions/concerns/issues you may have when getting started.